Tree UGM Demo
In the last demo we considered chain-structured data, one of the simplest types
of dependency where we can take advantage of the
graphical structure to allow efficient decoding/inference/sampling. In this
demo, we consider the case of tree-structured graphical models. In
particular, we consider the case where the edges in the graph can be
arbitrary, as long as the graph doesn't contain any loops. For models with this
type of dependency structure, we can still perform efficient
decoding/inference/sampling by applying generalizations of the methods
designed for chain-structured models.
Water Turbidity Problem
In some cities, it occasionally happens that snow melting off
of the mountain causes an increase in
the turbidity of the
drinking water at some locations. The turbidity level is used as a surrogate
for testing whether the water is safe to drink. For this reason, we may want
to build a probabilistic model of the water turbidity at different locations in
the water system. We will do this with a UGM, where we use a tree-structured
model to capture the dependencies between connected elements of the water system.
We will assume that turbidity is measured on a scale of 1 to 4, where 1
represents 'very safe', and 4 represents 'very unsafe'. We will assume that the water system can be
represented by the following graph:
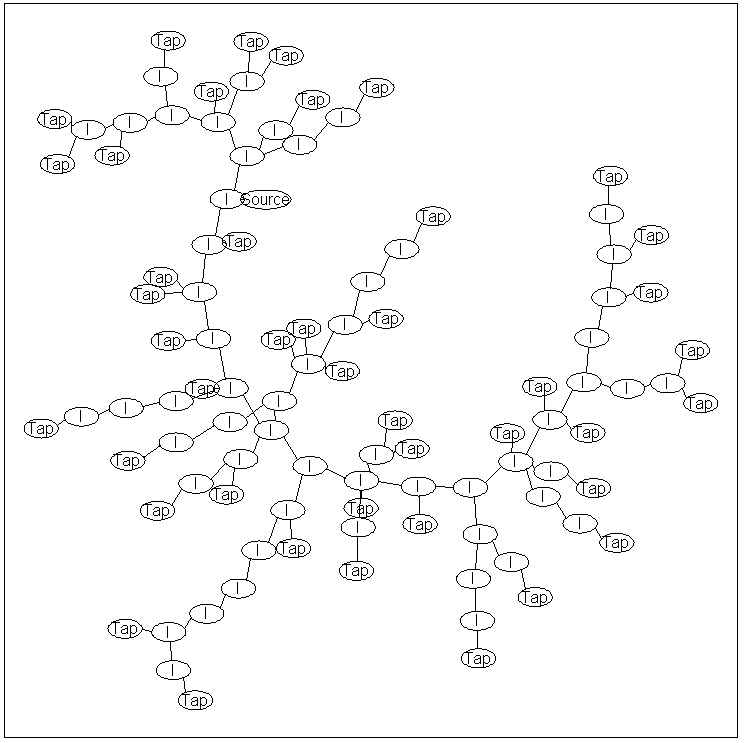
The node labeled 'Source' is the location where water enters the system.
Similarly, the nodes labeled 'Tap' are locations where water
exits the system, while nodes labeled 'I' are internal nodes.
The graph structure above is a 'tree', meaning that there are no loops in the graph and
the graph is connected (i.e., there exists a path of edges between all pairs of nodes).
We will assume that the node potentials for the source node are:
| State | Potential
|
| 1 | 0.9
|
| 2 | 0.09
|
| 3 | 0.009
|
| 4 | 0.001
|
That is, the water source will be 'very safe' 90% of the time, and 'very unsafe'
0.001% of the time. We will assign a unit value to all the other node potentials.
For the edge potentials that describe the dependencies between adjacent nodes, we
will use the following edge potential matrix:
| From/to | 1 | 2 | 3 | 4
|
| 1 | 0.9890 | 0.0099 | 0.0010 | 0.0001
|
| 2 | 0.1309 | 0.8618 | 0.0066 | 0.0007
|
| 3 | 0.0420 | 0.0841 | 0.8682 | 0.0057
|
| 4 | 0.0667 | 0.0333 | 0.1667 | 0.7333
|
In these edge potentials, the 'from' node represents the node that is closer to
the source. The model assumes that the turdity level will generally be the same between
adjacent nodes, with a small chance of it decreasing as we move away from the
source, and an even smaller chance of it increasing as we move away from the source.
Representing the water system with UGM
As before, we first make the adjacency matrix and edgeStruct. Rather than
typing out the large adjacency matrix, we will just load it from a .mat file:
load('waterSystem.mat'); % Loads adj
nNodes = length(adj);
nStates = 4;
edgeStruct = UGM_makeEdgeStruct(adj,nStates);
In the adjacency matrix, node 4 is the source node. We therefore make the
nodpePot matrix using:
source = 4;
nodePot = ones(nNodes,nStates);
nodePot(source,:) = [.9 .09 .009 .001];
Making the edgePot array is slightly more difficult, because we need to know
which end of the edge is closer to the source. Further, if the 2nd node is
closer to the source, we need to use the transpose of the edge potential matrix
above. We do this is in a fairly
simple (but not particularly efficient) way:
transition = [ 0.9890 0.0099 0.0010 0.0001
0.1309 0.8618 0.0066 0.0007
0.0420 0.0841 0.8682 0.0057
0.0667 0.0333 0.1667 0.7333];
colored = zeros(nNodes,1);
colored(source) = 1;
done = 0;
edgePot = zeros(nStates,nStates,edgeStruct.nEdges);
while ~done
done = 1;
colored_old = colored;
for e = 1:edgeStruct.nEdges
if sum(colored_old(edgeStruct.edgeEnds(e,:))) == 1
% Determine direction of edge and color nodes
if colored(edgeStruct.edgeEnds(e,1)) == 1
edgePot(:,:,e) = transition;
else
edgePot(:,:,e) = transition';
end
colored(edgeStruct.edgeEnds(e,:)) = 1;
done = 0;
end
end
end
Decoding
In this model, we have 100 nodes that each can take 4 possible states, so
we can't form a large table of possible states (with 4^100 elements) to do
decoding as we did in the first demo.
Further, our
dependency structure is no longer a chain, so we can no longer use the Viterbi
algorithm for decoding.
Fortunately, the dynamic programming ideas used in the Viterbi algorithm can be
extended to the case of a tree-structured dependency. This extension is often referred to
as
(max-product) belief propagation.
In the last demo, we mentioned that the forward-backward algorithms would give
the same result if we reversed the order of the nodes. That is, instead of
picking the last node as the 'end' and the first node as the 'start', we could
have made the first node the 'end' and the last node the 'start', and we would get the same result. Towards extending
this method to general trees, consider the following variation: we pick some
node in the middle of the chain, and define this to be the 'end'. We then pick
TWO 'start' nodes, the first node and the last node. We then run the forward
pass of the algorithm starting from both 'start' nodes until both of these
algorithms reach the 'end' node in the middle of the chain. We then combine
the 'messages' coming from both directions, compute the optimal value of the
'end' node in the middle of the chain, and then 'backtrack' in both directions
to get the optimal values for the rest of the chain.
It is straightforward to apply this idea to a general tree-structured graph:
- We define some arbitrary node in the graph as the 'end' (or 'root') node
- We define all nodes that have only one edge (excluding the
'root') as the 'start' nodes.
- We run the 'max' version of the 'forward' algorithm starting from each
'start', moving towards the 'root' and merging results when we reach
internal nodes.
- Once the results from all the algorithms have been merged at the root, we
backtrack through the tree to get the optimal decoding.
With UGM, we can run the max-product belief propagation algorithm on a
tree-structured model using:
optimalDecoding = UGM_Decode_Tree(nodePot,edgePot,edgeStruct)
In our model, the optimal decoding is that all nodes in the water system are 'very safe'.
Inference
We can make similar modifications to the forward-backward algorithm to get a
'sum-product' belief propagation algorithm that allows us to compute marginals
and the normalizing constant.
With UGM, we can run the sum-product belief propagation inference algorithm on a tree-structured model using:
[nodeBel,edgeBel,logZ] = UGM_Infer_Tree(nodePot,edgePot,edgeStruct)
nodeBel =
0.9103 0.0779 0.0110 0.0008
0.9110 0.0771 0.0111 0.0008
0.9129 0.0750 0.0114 0.0008
0.9000 0.0900 0.0090 0.0010
0.9073 0.0814 0.0104 0.0008
0.9129 0.0750 0.0114 0.0008
0.9116 0.0764 0.0112 0.0008
0.9085 0.0800 0.0106 0.0008
0.9043 0.0850 0.0099 0.0009
0.9085 0.0800 0.0106 0.0008
0.9129 0.0750 0.0114 0.0008
0.9121 0.0759 0.0112 0.0008
0.9141 0.0736 0.0115 0.0008
0.9110 0.0771 0.0111 0.0008
0.9085 0.0800 0.0106 0.0008
0.9059 0.0830 0.0102 0.0009
0.9073 0.0814 0.0104 0.0008
0.9103 0.0779 0.0110 0.0008
0.9125 0.0754 0.0113 0.0008
0.9116 0.0764 0.0112 0.0008
0.9138 0.0739 0.0115 0.0008
0.9125 0.0754 0.0113 0.0008
0.9085 0.0800 0.0106 0.0008
0.9103 0.0779 0.0110 0.0008
0.9095 0.0789 0.0108 0.0008
0.9132 0.0746 0.0114 0.0008
0.9129 0.0750 0.0114 0.0008
0.9129 0.0750 0.0114 0.0008
0.9116 0.0764 0.0112 0.0008
0.9129 0.0750 0.0114 0.0008
0.9116 0.0764 0.0112 0.0008
0.9110 0.0771 0.0111 0.0008
0.9116 0.0764 0.0112 0.0008
0.9121 0.0759 0.0112 0.0008
0.9140 0.0737 0.0115 0.0008
0.9132 0.0746 0.0114 0.0008
0.9121 0.0759 0.0112 0.0008
0.9073 0.0814 0.0104 0.0008
0.9116 0.0764 0.0112 0.0008
0.9116 0.0764 0.0112 0.0008
0.9134 0.0743 0.0114 0.0008
0.9073 0.0814 0.0104 0.0008
0.9134 0.0743 0.0114 0.0008
0.9095 0.0789 0.0108 0.0008
0.9138 0.0739 0.0115 0.0008
0.9125 0.0754 0.0113 0.0008
0.9132 0.0746 0.0114 0.0008
0.9116 0.0764 0.0112 0.0008
0.9095 0.0789 0.0108 0.0008
0.9134 0.0743 0.0114 0.0008
0.9073 0.0814 0.0104 0.0008
0.9059 0.0830 0.0102 0.0009
0.9138 0.0739 0.0115 0.0008
0.9116 0.0764 0.0112 0.0008
0.9132 0.0746 0.0114 0.0008
0.9140 0.0737 0.0115 0.0008
0.9059 0.0830 0.0102 0.0009
0.9103 0.0779 0.0110 0.0008
0.9132 0.0746 0.0114 0.0008
0.9132 0.0746 0.0114 0.0008
0.9110 0.0771 0.0111 0.0008
0.9095 0.0789 0.0108 0.0008
0.9132 0.0746 0.0114 0.0008
0.9073 0.0814 0.0104 0.0008
0.9137 0.0741 0.0115 0.0008
0.9116 0.0764 0.0112 0.0008
0.9059 0.0830 0.0102 0.0009
0.9134 0.0743 0.0114 0.0008
0.9138 0.0739 0.0115 0.0008
0.9023 0.0873 0.0095 0.0009
0.9116 0.0764 0.0112 0.0008
0.9103 0.0779 0.0110 0.0008
0.9125 0.0754 0.0113 0.0008
0.9121 0.0759 0.0112 0.0008
0.9085 0.0800 0.0106 0.0008
0.9073 0.0814 0.0104 0.0008
0.9116 0.0764 0.0112 0.0008
0.9121 0.0759 0.0112 0.0008
0.9095 0.0789 0.0108 0.0008
0.9134 0.0743 0.0114 0.0008
0.9137 0.0741 0.0115 0.0008
0.9116 0.0764 0.0112 0.0008
0.9110 0.0771 0.0111 0.0008
0.9129 0.0750 0.0114 0.0008
0.9121 0.0759 0.0112 0.0008
0.9059 0.0830 0.0102 0.0009
0.9085 0.0800 0.0106 0.0008
0.9095 0.0789 0.0108 0.0008
0.9132 0.0746 0.0114 0.0008
0.9121 0.0759 0.0112 0.0008
0.9085 0.0800 0.0106 0.0008
0.9129 0.0750 0.0114 0.0008
0.9110 0.0771 0.0111 0.0008
0.9103 0.0779 0.0110 0.0008
0.9110 0.0771 0.0111 0.0008
0.9073 0.0814 0.0104 0.0008
0.9043 0.0850 0.0099 0.0009
0.9132 0.0746 0.0114 0.0008
0.9121 0.0759 0.0112 0.0008
0.9121 0.0759 0.0112 0.0008
We see in row 4 that the source node's marginals match its node potentials. We
also see that the probability of the water being 'very safe' is higher than the
source's maginals for all other nodes, while the probability of the other levels
is decreased.
In this specific model, the node potentials of the source node can be
interpreted as a marginal probability and the values of the edge potentials can
be interpreted as transition probabilities. Further, it is possible to use simpler forward
inference and sampling methods, similar to the Chapman-Kolmogorov equations
for Markov chains (in this case, we apply the analogous equations to an
appropriately
defined branching
process, where we start at the source, branch at internal nodes, and
terminate at tap nodes).
This probabilistic interpretation will not hold for general tree-structured
UGMs. However, the methods implemented in UGM handle the
case of general tree-structured UGMs, and can therefore handle the extensions
to non-unit potentials (beyond the source node), or edge potentials that
aren't normalized probabilities as we move away from the source.
Sampling
Sampling from a tree-structured UGM is again a straightforward extension of the
forwards-filter backwards-sample algorithm for chain-structured UGMs.
With UGM, we can generate 100 samples from a tree-structured model using:
edgeStruct.maxIter = 100;
samples = UGM_Sample_Tree(nodePot,edgePot,edgeStruct);
Here is a visualization of 100 samples:
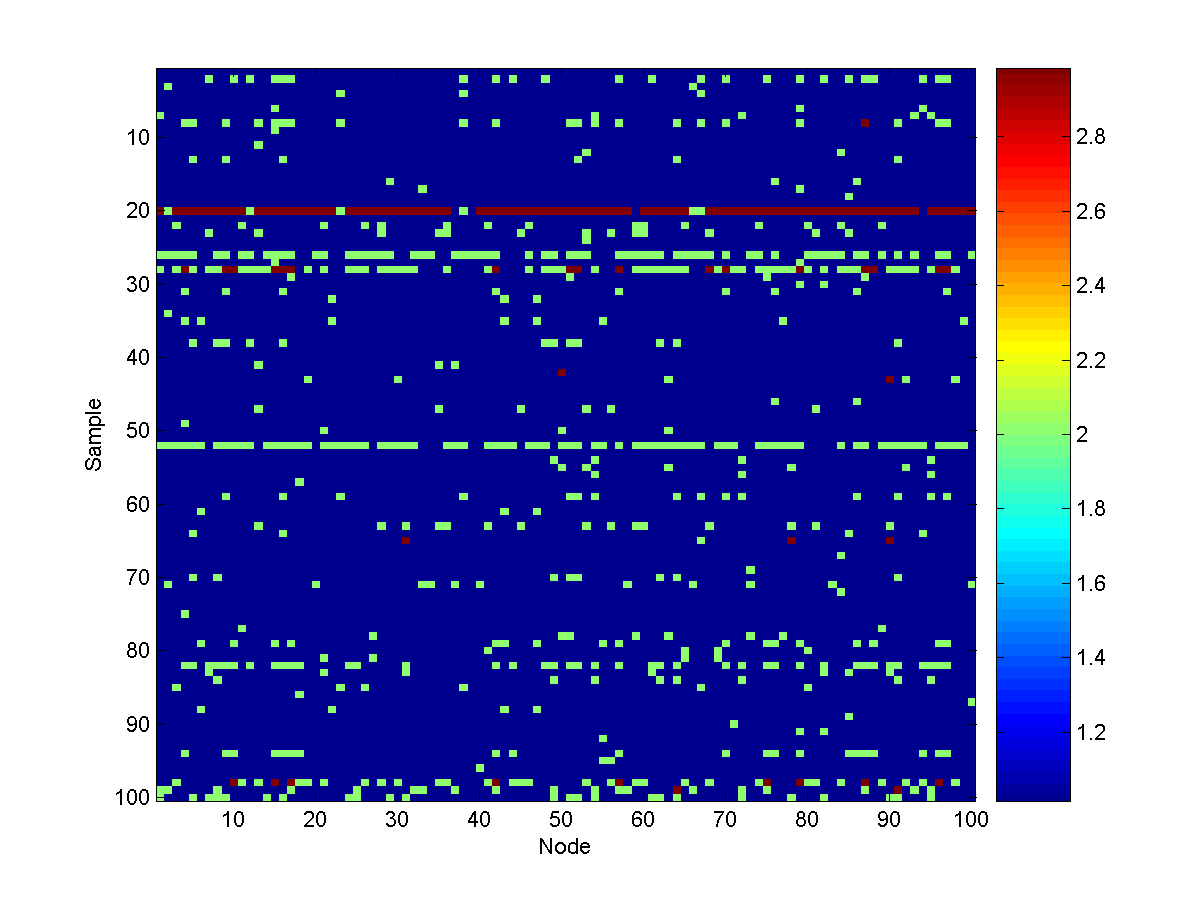
From these samples, we see that turbidity can spread widely throughout the
water system, depending on how far the source is from the 'very safe' state.
Notes
Since chains are a special case of trees, for the last two demos we could have used the tree
decoding/inference/sampling methods discussed in this demo. Further, the tree
decoding/inference/sampling methods implemented in UGM can handle the case of
'forests'. A forest-structured graph is a graph that has no loops. This generalizes tree-structured
graphs, because the forests do not need to be connected. Said another way, a forest
is a union of non-overlapping tree-structured graphs.
In the samples generated above, the state '4' was
never encountered. Although we would eventually generate samples where state 4
was encountered, knowing how the model behaves in this scenario is clearly
important and we may not want to wait for these unlikely scenarios to
occur. In the next demo, we examine performing conditional
decoding/inference/sampling, where we perform these operations under the
assumption that the values of one or more nodes are known (ie. what
happens to the other nodes when the source node is in state 4).
PREVIOUS DEMO NEXT DEMO
Mark Schmidt > Software > UGM > Tree Demo

