Junction UGM Demo
For performing exact decoding/inference/sampling in loopy UGMs, the main alternative to cutset conditioning is the junction tree method. Roughly, this method aggregates sets of nodes into larger supernodes that form a tree structure. It then performs exact inference on the tree structure. This will be efficient if the supernodes do not have to be too big in order to make the structure into a tree.
Plane Infection Model
People often get sick from travelling by commercial airplanes. One of the key factors affecting whether you will get sick is how close you sit to an infected person. We will build a simple model of the spread of infections on airplanes.
As a simple model of an airplane seating pattern, we will use the following lattice-structured graph:
( 1)-( 2)-( 3)-( 4)-( 5)-( 6)
| | | | | |
( 7)-( 8)-( 9)-( 10)-( 11)-( 12)
| | | | | |
( 13)-( 14)-( 15)-( 16)-( 17)-( 18)
| | | | | |
( 19)-( 20)-( 21)-( 22)-( 23)-( 24)
| | | | | |
( 25)-( 26)-( 27)-( 28)-( 29)-( 30)
| | | | | |
( 31)-( 32)-( 33)-( 34)-( 35)-( 36)
| | | | | |
( 37)-( 38)-( 39)-( 40)-( 41)-( 42)
| | | | | |
( 43)-( 44)-( 45)-( 46)-( 47)-( 48)
| | | | | |
( 49)-( 50)-( 51)-( 52)-( 53)-( 54)
| | | | | |
( 55)-( 56)-( 57)-( 58)-( 59)-( 60)
| | | | | |
( 61)-( 62)-( 63)-( 64)-( 65)-( 66)
| | | | | |
( 67)-( 68)-( 69)-( 70)-( 71)-( 72)
| | | | | |
( 73)-( 74)-( 75)-( 76)-( 77)-( 78)
| | | | | |
( 79)-( 80)-( 81)-( 82)-( 83)-( 84)
| | | | | |
( 85)-( 86)-( 87)-( 88)-( 89)-( 90)
| | | | | |
( 91)-( 92)-( 93)-( 94)-( 95)-( 96)
| | | | | |
( 97)-( 98)-( 99)-(100)-(101)-(102)
| | | | | |
(103)-(104)-(105)-(106)-(107)-(108)
| | | | | |
(109)-(110)-(111)-(112)-(113)-(114)
| | | | | |
(115)-(116)-(117)-(118)-(119)-(120)
| | | | | |
(121)-(122)-(123)-(124)-(125)-(126)
| | | | | |
(127)-(128)-(129)-(130)-(131)-(132)
| | | | | |
(133)-(134)-(135)-(136)-(137)-(138)
| | | | | |
(139)-(140)-(141)-(142)-(143)-(144)
| | | | | |
(145)-(146)-(147)-(148)-(149)-(150)
| | | | | |
(151)-(152)-(153)-(154)-(155)-(156)
| | | | | |
(157)-(158)-(159)-(160)-(161)-(162)
| | | | | |
(163)-(164)-(165)-(166)-(167)-(168)
| | | | | |
(169)-(170)-(171)-(172)-(173)-(174)
| | | | | |
(175)-(176)-(177)-(178)-(179)-(180)
| | | | | |
(181)-(182)-(183)-(184)-(185)-(186)
| | | | | |
(187)-(188)-(189)-(190)-(191)-(192)
| | | | | |
(193)-(194)-(195)-(196)-(197)-(198)
| | | | | |
(199)-(200)-(201)-(202)-(203)-(204)
| | | | | |
(205)-(206)-(207)-(208)-(209)-(210)
| | | | | |
(211)-(212)-(213)-(214)-(215)-(216)
| | | | | |
(217)-(218)-(219)-(220)-(221)-(222)
| | | | | |
(223)-(224)-(225)-(226)-(227)-(228)
| | | | | |
(229)-(230)-(231)-(232)-(233)-(234)
| | | | | |
(235)-(236)-(237)-(238)-(239)-(240)
We will set the node potentials to give a preference towards not being infected, and use the edge potentials to reflect that adjacent passengers are more likely to be co-infected.
Representing the plane infection model with UGM
We first make the adjacency matrix and the edgeStruct:
nSeats = 6;
nRows = 40;
nNodes = nSeats*nRows;
nStates = 2;
adj = zeros(nNodes);
for r = 1:nRows
for s = 1:nSeats
if s < nSeats
adj(s + nSeats*(r-1),(s+1) + nSeats*(r-1)) = 1; % Passenger behind
end
if r < nRows
adj(s + nSeats*(r-1),s + nSeats*(r)) = 1; % Passenger to the right
end
end
end
adj = adj+adj'; % Symmetrize
edgeStruct = UGM_makeEdgeStruct(adj,nStates);
And then the potentials:
alpha = .5;
beta = 2;
nodePot = [ones(nNodes,1) alpha*ones(nNodes,1)];
edgePot = repmat([beta 1;1 beta],[1 1 edgeStruct.nEdges]);
We use the parameter alpha to control the baseline rate of infection, and beta to control how effectively the infection spreads.
Exact Decoding/Inference/Sampling
Despite the large number of loops in the graph and the lack of an obvious cutset, the graph structure only a has a tree width of 6. This allows us to use the junction tree algorithm for exact decoding/inference/sampling. Roughly, the junction tree aggregates together the passengers seated in the same row of the plane to create a single super-node. It then passes messages between these super-nodes, which form a chain-structured graph.
To do exact decoding/inference/sampling in this model with the junction tree algorithm, we can use:
optimalDecoding = UGM_Decode_Junction(nodePot,edgePot,edgeStruct);
[nodeBel,edgeBel,logZ] = UGM_Infer_Junction(nodePot,edgePot,edgeStruct);
samples = UGM_Sample_Junction(nodePot,edgePot,edgeStruct);
We can draw individual samples from the model as an image. Below are four examples:
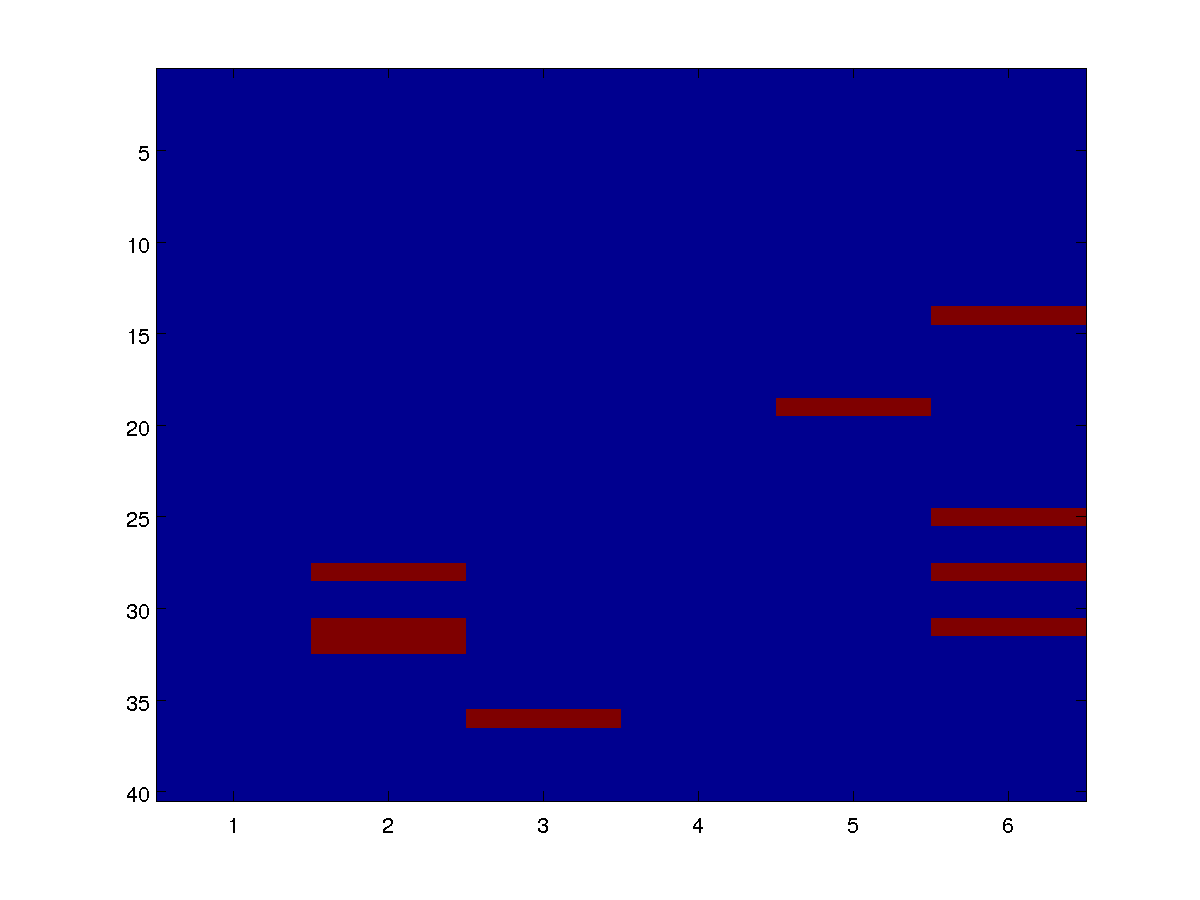
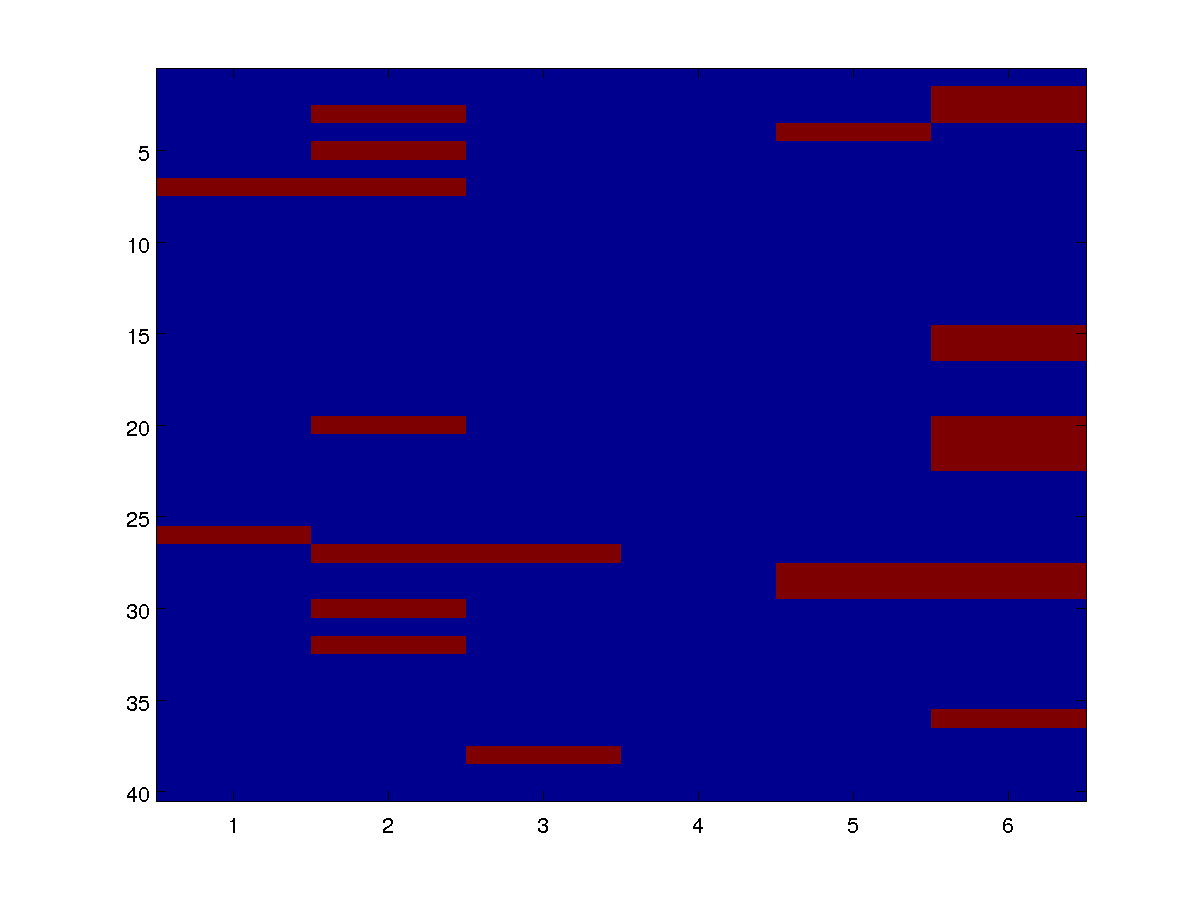
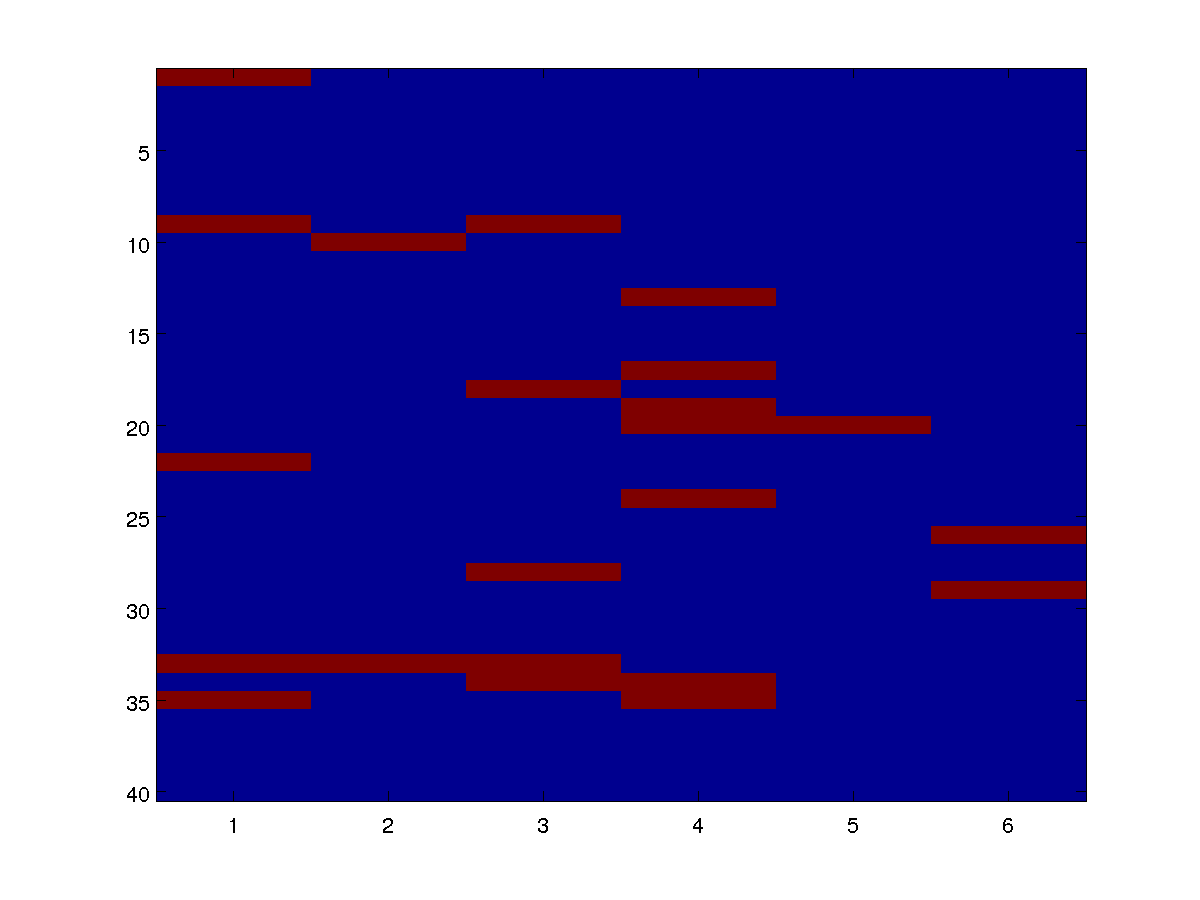
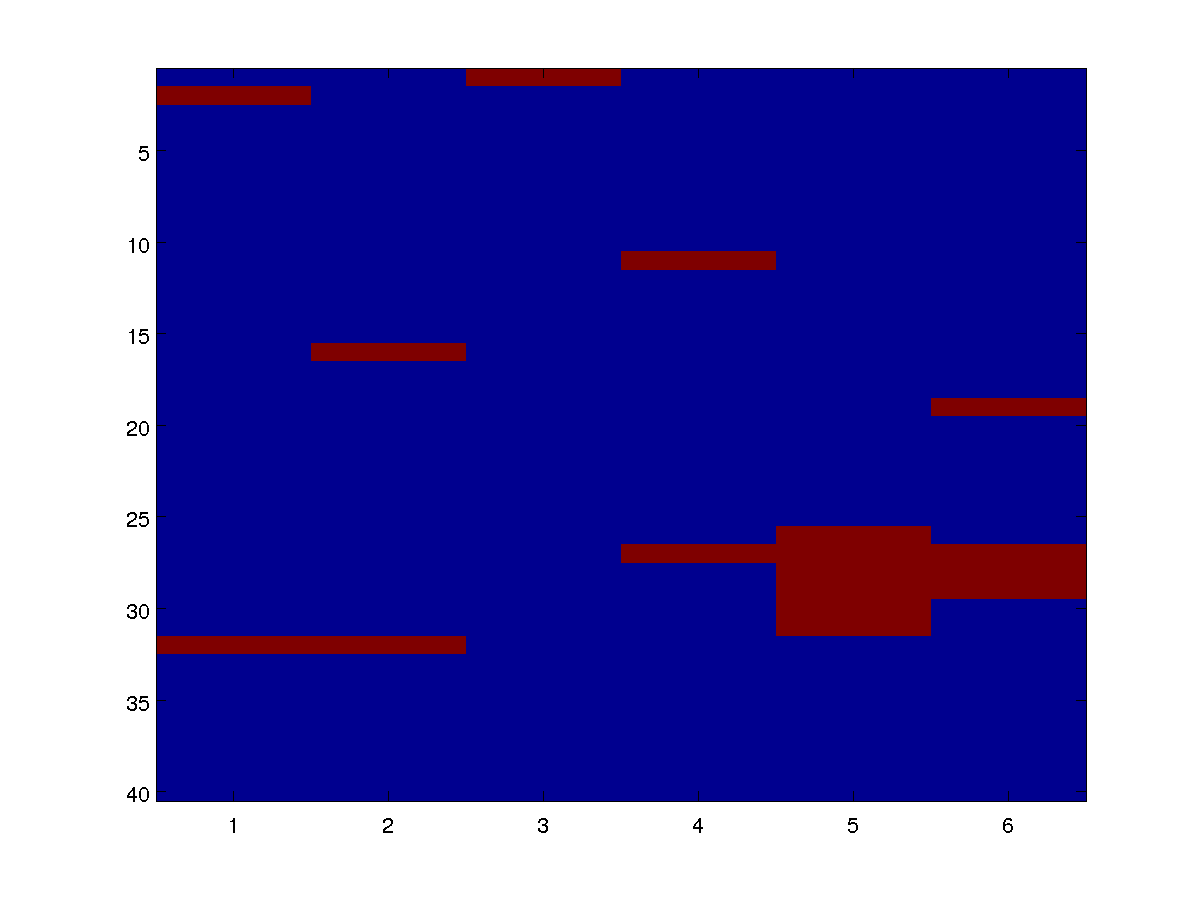
By varying the parameters alpha and beta in the definitions of the potentials, we can see how the distribution changes based on the base rate and the infectiousness.
Notes
Be default, the junction tree codes in UGM assume the ordering 1:nNodes. This may lead to a sub-optimal junction tree, and an alternate ordering can be passed to these functions as a fourth argument.
The junction tree methods can also be used for efficient exact decoding/inference/sampling in the all of the models discussed in previous demos. However, note that my junction tree implementation is not particularly efficient. The airplane infection model was suggested by Martin Wainwright in his lecture at MLSS'11.
I'm not confident that my junction tree sampling code is correct. The clique marginals seem to be computed correctly, but the composition sampler implementing the backwards-sampling phase seems to have a bug. Right now I don't have the time/patience to find/fix this bug, but let me know if you find/fix it!
PREVIOUS DEMO NEXT DEMO
Mark Schmidt > Software > UGM > Junction Demo



