Aggregated Dendrograms for Visual Comparison Between Many Phylogenetic Trees
IEEE Transactions on Visualization and Computer
Graphics (TVCG), 29(9):2732-2747 2019.
DOI: 10.1109/TVCG.2019.2898186
DOI: 10.1109/TVCG.2019.2898186
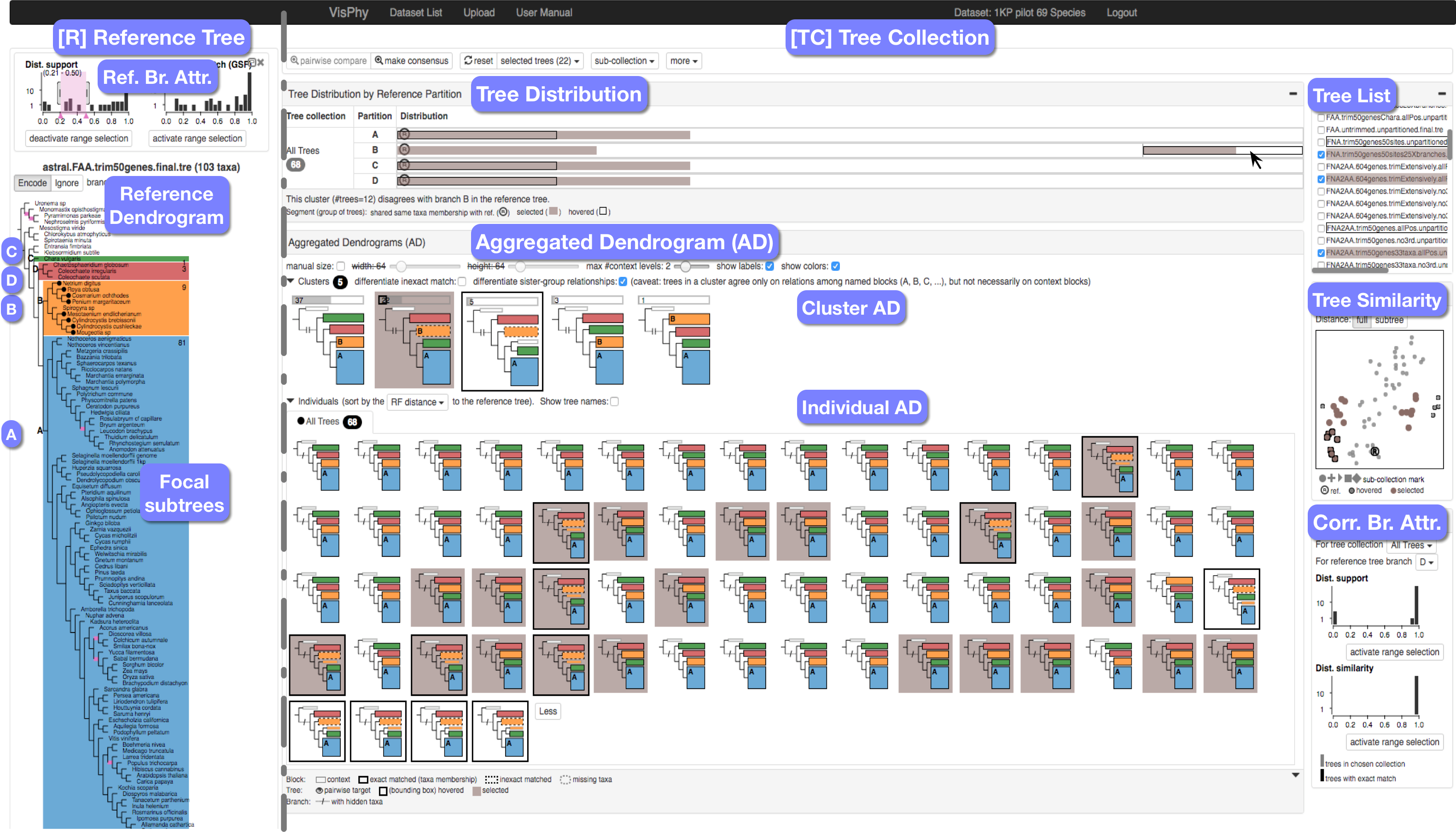
Fig 1. Screenshot of ADView comparing a reference tree (left pane) against a tree collection (right pane).
Abstract
We address the visual comparison of multiple phylogenetic trees that arises in evolutionary biology, specifically between one reference tree and a collection of dozens to hundreds of other trees. We abstract the domain questions of phylogenetic tree comparison
as tasks to look for supporting or conflicting evidence for hypotheses that requires inspection of both topological structure and attribute values at different levels of detail in the tree collection. We introduce the new visual encoding
idiom of aggregated dendrograms to concisely summarize the topological relationships between interactively chosen focal subtrees according to biologically meaningful criteria, and provide a layout algorithm that automatically adapts
to the available screen space. We design and implement the ADView system, which represents trees at multiple levels of detail across multiple views: the entire collection, a subset of trees, an individual tree, specific subtrees of
interest, and the individual branch level. We benchmark the algorithms developed for ADView, compare its information density to previous work, and demonstrate its utility for quickly gathering evidence about biological hypotheses through
usage scenarios with data from recently published phylogenetic analysis and case studies of expert use with real-world data, drawn from a summative interview study.
Materials
- Paper: preprint| IEEE Digital Library| bib
- Videos: demo video (4min)| high resolution version on youtube
- Supplemental: pdf
- Demo: link
- Code: frontend| backend
- Talk (at InfoVis 2019, Vancouver): slides (pptx)| video on Vimeo
- Figures: high-res