SequenceJuxtaposer: Fluid Navigation For Large-Scale Sequence
Comparison In Context
James Slack, Kristian Hildebrand, Tamara Munzner, and Katherine St. John
Proc. German Conference on Bioinformatics 2004, pp 37-42
PDF |
Abstract |
Videos |
Software |
Figures |
Talk
Paper
Abstract
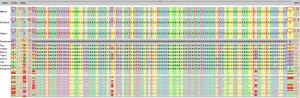
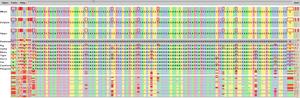
SequenceJuxtaposer is a sequence visualization tool for the
exploration and comparison of biomolecular sequences. We use an
information visualization technique called ``accordion drawing'' that
guarantees three key properties: context, visibility, and frame rate.
We provide context through the navigation metaphor of a rubber sheet
that can be smoothly stretched to show more details in the areas of
focus, while the surrounding regions of context are correspondingly
shrunk. Landmarks, such as user specified motifs or
differences between aligned base pairs across multiple sequences, are
guaranteed to be visible even if located in the shrunken areas of
context. Our graphics infrastructure for progressive rendering
provides immediate responsiveness to user interaction by guaranteeing
that we redraw the scene at a target frame rate. Our preprocessing
algorithms are subquadratic: O(nk) for k sequences of n base
pairs each. All runtime rendering algorithms are sublinear in nk: they are
O(v) where v is the number of items visible onscreen at once,
and v \ll nk. SequenceJuxtaposer supports interaction at 20 frames
per second when browsing collections of several hundred sequences that
comprise over 1.7 million total base pairs.
We show three example applications with large, publicly available
datasets, and we are able to quickly observe many features that had
previously required significant analysis to discover.
Videos
QuickTime videos include voiceover
Software
Figures
Talk
PPT, HTML, PDF
1up,
6up
Tamara Munzner
Last modified: Sun Nov 20 15:52:22 PST 2005