Selecting Lambda Using the ModelSelection Class
In this example, we demonstrate how to use the ModelSelection class to choose the L2 regularization parameter lambda for a LinregDist model. We use four different scoring functions: CV (MSE loss), CV (NLL loss), BIC, AIC.
Contents
We will use the prostate data set and create our base model, the LinregDist object, which will be used throughout.
load prostate; verbose = true; T = ChainTransformer({StandardizeTransformer(false),AddOnesTransformer()}); baseModel = LinregDist('transformer',T);
Model Space
In this example, we will use the built in exhaustive search function and so we must specify the full range of lambdas we will search over.
When using the model selection class, models are represented as a collection of values. Any valid Matlab data type can be used including objects, doubles, strings etc. Each model is stored as a cell array and the models are stacked vertically. In particular, models{1} returns the first model and models{1}{1} returns the first element of the first model. By using this representation, we can, in general, select from among models with differing numbers of parameters.
We can use the static method formatModels() to help create our model space. We simply pass in the range we wish to search over and it returns the model space in the appropriate format.
models = ModelSelection.makeModelSpace(logspace(-5,3,50));
We will see in the 2d example that an n-dimensional model space can be created by simply passing in a range for each dimensions, e.g.
models = ModelSelection.makeModelSpace(0:0.1:1, 1:10)
Cross Validation (MSE loss)
The simplest case uses cross validation as our scoring function with mean squared error loss. While custom scoring functions can be used with the class, the cross validation function is built in, as is the mse loss function.
Test Function
We must specify a test function, which will be called by the cross validation scoring function each fold for each given lambda. This function must train the LinregDist on the specified data using the given lambda and return the predicted target values. These will be passed directly to the built in mse loss function. Here, and in general, the test function will be the composition of several other functions. In this case we use fit, predict, and mode.
predictFunction = @(Xtrain,ytrain,Xtest,lambda)... mode(predict(fit(baseModel,'X',Xtrain,'y',ytrain,'prior','l2','lambda',lambda,'prior','l2'),Xtest));
Run Model Selection
To perform the model selection, we simply call the ModelSelection constructor with the right inputs. Unless we turn it off with "'doPlot',false" , a plot is automatically generated which we can further customize using the usual commands. The selected model is stored in the ModelSelection object; we use this to retrain the model on all of the data and assess the performance.
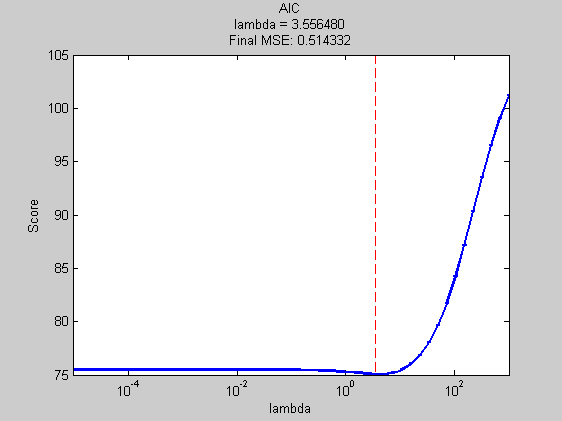
msCVmse = ModelSelection( ... 'predictFunction',predictFunction ,... % the test function we just created 'Xdata' ,Xtrain ,... % all of the X data we have available 'Ydata' ,ytrain ,... % all of the y data we have available 'verbose' ,verbose ,... % turn off progress report 'models' ,models ); % the model space created above lambda = msCVmse.bestModel{1}; % assess performance of chosen lambda cvFinalError = mse(ytest,predictFunction(Xtrain,ytrain,Xtest,lambda)); title(sprintf('CV mse loss\nlambda = %f\nFinal MSE: %f',msCVmse.bestModel{1},cvFinalError)); xlabel('lambda');
Best Model = [2.44]
Cross Validation (NLL Loss)
We now perform cross validation but use negative log likelihood as our loss function. Recall that the output of our predictFunction is passed directly to the loss function. In this case we will have the predictFunction simply return the fitted model. We will then define a custom loss function to calculate the nll.
predictFunction = @(Xtrain,ytrain,Xtest,lambda)... fit(baseModel,'X',Xtrain,'y',ytrain,'lambda',lambda,'prior','l2');
Loss Function
When defining our own scoring function as we will do later, we are free to define the test and loss functions any way we wish, however to use the built in CV function, our loss function must take in three arguments. the first should be the output from the test function
lossFunction = @(fittedObj,Xtest,ytest)-logprob(fittedObj,Xtest,ytest);
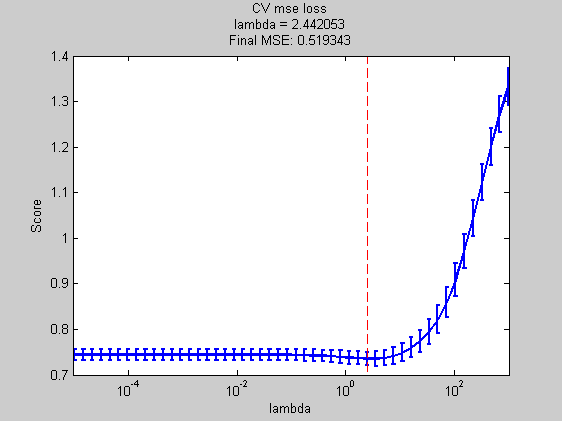
msCVnll = ModelSelection( ... 'predictFunction' ,predictFunction ,... 'lossFunction' ,lossFunction ,... 'Xdata' ,Xtrain ,... 'Ydata' ,ytrain ,... 'verbose' ,verbose ,... 'models' ,models ); cvnllFinalError = mse(ytest,mode(predict(... fit(baseModel,'X',Xtrain,'y',ytrain,'lambda',msCVnll.bestModel{1}),Xtest))); title(sprintf('CV NLL\nlambda = %f\nFinal MSE: %f',msCVnll.bestModel{1},cvnllFinalError)); xlabel('lambda');
Best Model = [16.00]
BIC
To perform model selection using BIC, we must define a custom scoring function. If we use the built in exhaustive search, this function must take two parameters: (1) the ModelSelection object and (2) a candidate model stored as a cell array. Our custom scoring function is free to access any of the public properties of the ModelSelection class and use the test and loss functions any way it likes, (assuming we have defined them). In this case, however, we will not need these functions and simply redefine the scoring function.
Scoring Function
We use the bicScore function of the LinregDist class to do most of the work. We must first fit the base LinregDist object using the current lambda value, which is passed by the search function in model{1}.
scoreFcn = @(obj,model)... -bicScore(... fit(baseModel,'X',Xtrain,'y',ytrain,'lambda',model{1},'prior','l2')... ,Xtrain,ytrain,model{1});
We now run the model selection.
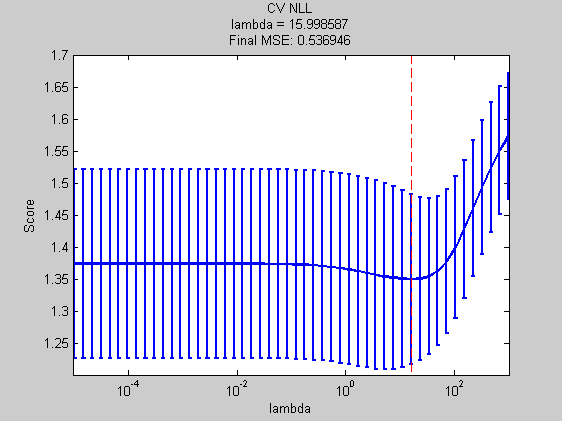
msBic = ModelSelection( ... 'Xdata' ,Xtrain ,... 'Ydata' ,ytrain ,... 'models' ,models ,... 'verbose' ,verbose ,... 'scoreFunction' ,scoreFcn ); bicFinalError = mse(ytest,mode(predict(... fit(baseModel,'X',Xtrain,'y',ytrain,'prior','l2','lambda',msBic.bestModel{1}),Xtest))); title(sprintf('BIC\nlambda = %f\nFinal MSE: %f',msBic.bestModel{1},bicFinalError)); xlabel('lambda');
Best Model = [16.00]
AIC
The AIC case is almost identical to the BIC case.
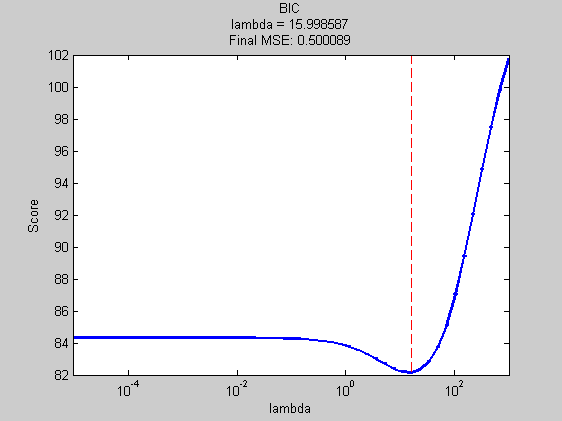
scoreFcn = @(obj,model) -aicScore(fit(baseModel,'X',Xtrain,'y',ytrain,'prior','l2','lambda',model{1}),Xtrain,ytrain,model{1}); msAic = ModelSelection( ... 'Xdata' ,Xtrain ,... 'Ydata' ,ytrain ,... 'models' ,models ,... 'verbose' ,verbose ,... 'scoreFunction' ,scoreFcn ); aicFinalError = mse(ytest,mode(predict(... fit(baseModel,'X',Xtrain,'y',ytrain,'prior','l2','lambda',msAic.bestModel{1}),Xtest))); title(sprintf('AIC\nlambda = %f\nFinal MSE: %f',msAic.bestModel{1},aicFinalError)); xlabel('lambda');
Best Model = [3.56]