 For a non-technical introduction to Bayesian networks, read
this LA times article
(10/28/96).
For some of the technical details,
see my tutorial below.
For the really gory details,
see the AUAI homepage
(Association for Uncertainty in Artificial Intelligence), or my list
of recommended reading at the end.
The UAI mailing list
is now archived and provides up-to-date information.
I also maintain a list of Bayes Net software
packages, including my own Bayes Net
Toolbox for Matlab.
Note that, despite the name,
Bayesian networks do not necessarily imply a commitment to Bayesian
methods. Rather, they are so called because they use Bayes' rule for
probailistic inference (see below).
For non-technical introductions to Bayesian methods,
see this Economist article (9/30/00)
or this New York Times article (4/28/01).
For more technical details,
see
Tom Minka's excellent tutorial notes.
For a non-technical introduction to Bayesian networks, read
this LA times article
(10/28/96).
For some of the technical details,
see my tutorial below.
For the really gory details,
see the AUAI homepage
(Association for Uncertainty in Artificial Intelligence), or my list
of recommended reading at the end.
The UAI mailing list
is now archived and provides up-to-date information.
I also maintain a list of Bayes Net software
packages, including my own Bayes Net
Toolbox for Matlab.
Note that, despite the name,
Bayesian networks do not necessarily imply a commitment to Bayesian
methods. Rather, they are so called because they use Bayes' rule for
probailistic inference (see below).
For non-technical introductions to Bayesian methods,
see this Economist article (9/30/00)
or this New York Times article (4/28/01).
For more technical details,
see
Tom Minka's excellent tutorial notes.
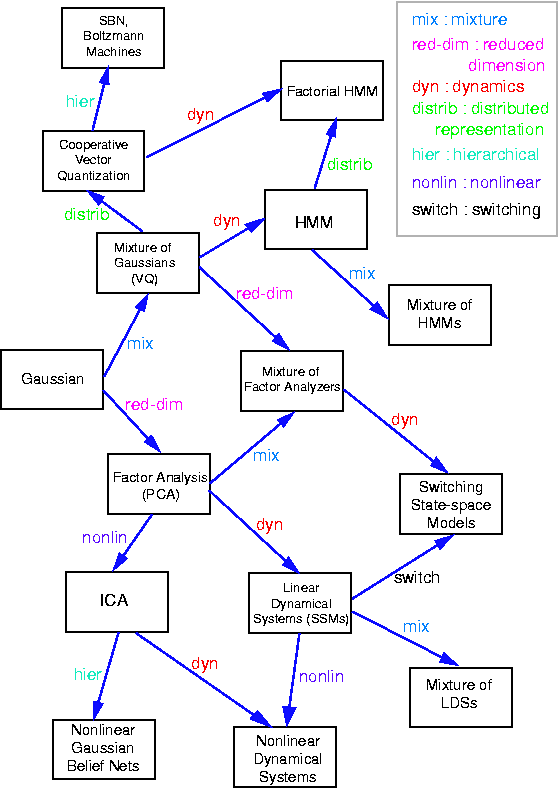
"Graphical models are a marriage between probability theory and
graph theory. They provide a natural tool for dealing with two problems
that occur throughout applied mathematics and engineering --
uncertainty and complexity -- and in particular they are playing an
increasingly important role in the design and analysis of machine
learning algorithms. Fundamental to the idea of a graphical model is
the notion of modularity -- a complex system is built by combining
simpler parts. Probability theory provides the glue whereby the parts
are combined, ensuring that the system as a whole is consistent, and
providing ways to interface models to data. The graph theoretic side
of graphical models provides both an intuitively appealing interface
by which humans can model highly-interacting sets of variables as well
as a data structure that lends itself naturally to the design of
efficient general-purpose algorithms.
Many of the classical multivariate probabalistic systems studied in
fields such as statistics, systems engineering, information theory,
pattern recognition and statistical mechanics are special cases of the
general graphical model formalism -- examples include mixture models,
factor analysis, hidden Markov models, Kalman filters and Ising
models. The graphical model framework provides a way to view all of
these systems as instances of a common underlying formalism. This view
has many advantages -- in particular, specialized techniques that have
been developed in one field can be transferred between research
communities and exploited more widely. Moreover, the graphical model
formalism provides a natural framework for the design of new systems."
We will briefly discuss the following topics.
There are two main kinds of graphical models: undirected and directed. Undirected graphical models, also known as Markov networks or Markov random fields (MRFs), are more popular with the physics and vision communities. (Log-linear models are a special case of undirected graphical models, and are popular in statistics.) Directed graphical models, also known as generative models or Bayesian/ belief networks (BNs), are more popular with the AI and machine learning communities. (Note that, despite the name, Bayesian networks do not necessarily imply a commitment to Bayesian methods; rather, they are so called because they use Bayes' rule for inference, as we will see below.) It is also possible to have a model with both directed and undirected arcs, which is called a chain graph.

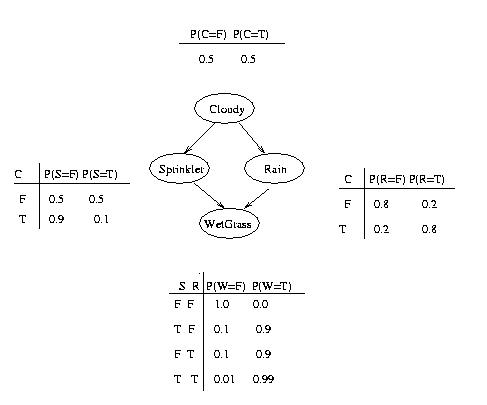
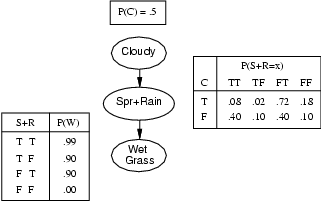
Consider the example above. Here, nodes represent binary random variables. We see that the event ``grass is wet'' (W=true) has two possible causes: either the water sprinker is on (S=true) or it is raining (R=true). The strength of this relationship is shown in the table below W; this is called W's conditional probability table (CPT). For example, we see that P(W=true | S=true, R=false) = 0.9 (second entry of second row), and hence, P(W=false | S=true, R=false) = 1 - 0.9 = 0.1, since each row must sum to one. Since the C node has no parents, its CPT specifies the prior probability that it is cloudy (in this case, 0.5).
The simplest statement of the conditional independence relationships encoded in a Bayesian network can be stated as follows: a node is independent of its ancestors given its parents, where the ancestor/parent relationship is with respect to some fixed topological ordering of the nodes. Let us see how we can use this fact to specify the joint distribution more compactly.
By the chain rule of probability, the joint probability of all the nodes in the graph above is
P(C, S, R, W) = P(C) * P(S|C) * P(R|C,S) * P(W|C,S,R)By using conditional independence relationships, we can rewrite this as
P(C, S, R, W) = P(C) * P(S|C) * P(R|C) * P(W|S,R)where we were allowed to simplify the third term because R is independent of S given its parent C (written R \indep S | C), and the last term because W \indep C | S, R.
We can see that the conditional independence relationships allow us to represent the joint more compactly. Here the savings are minimal, but in general, if we had n binary nodes, the full joint would require O(2^n) parameters, but the factored form would only require O(n 2^k) parameters, where k is the maximum fan-in of a node.
We will now show how many popular statistical models can be represented as directed graphical models.
| 
|
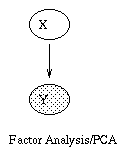
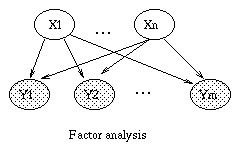
If we let the prior distribution on X be P(X=x) = N(x; 0, I), and the conditional distribution on Y be P(Y=y|X=x) = N(y; \mu + W x, \Psi), where \Psi is a diagonal matrix, then this model is called factor analysis. (The notation N(x; \mu, \Sigma) means the value of a Gaussian pdf with mean \mu and covariance \Sigma evaluated at the point (vector) x.) We usually assume that n \ll m, where X \in \real^n and Y \in \real^m, so the model is trying to explain a high dimensional vector as a linear combination of low dimensional features.
A common simplification is to assume isotropic noise, i.e., \Psi = \sigma I, for some scalar \sigma. In this case, the components of the vectors X and Y are uncorrelated, which we can explicitely represent using the model in Figure (b). It turns out that the maximum likelihood estimate of the factor loading matrix W in this case is given by the first m principle eigenvectors of the sample covariance matrix, with scalings determined by the eigenvalues and sigma. Classical PCA can be obtained by taking the \sigma \rightarrow 0 limit \cite{Tipping99,Roweis99}.
| 
|
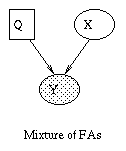
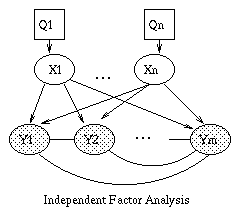
We can lift the restrictive assumption of linearity by modelling Y as a {\em mixture} of linear subspaces \cite{Tipping99}. This model is is shown in Figure~\ref{fig:mfa}(a). Q is a discrete latent (hidden) variable, that indicates which of the subspaces to use to generate Y. (In this tutorial, we adopt the (non-standard) convention notation that square nodes represent discrete random variables, and circles represent continuous random variables. We also use the (standard) convention that shaded nodes are observed, and clear nodes are (usually) hidden.) Mathematically, this can be written as
P(Q=i) &=& \pi_i \\ P(X=x) &=& N(x; 0, I) \\ P(Y=y|X=x,Q=i) &=& N(y; \mu_i + W_i x, \Psi_i)
Similarly, we can lift the restrictive assumption of Gaussian noise by modelling the prior on X as a mixture of Gaussians. One way to do this, known as independent factor analysis (IFA) \cite{Attias98}, is shown in Figure (b). (Technically, this is a chain graph: the undirected arcs between the components of Y indicate that they are correlated, i.e., that \Psi is not diagonal). This model is closely related to independent components analysis (ICA).
For more details, see the following papers.
| 
|
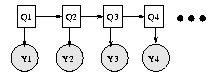
Consider the Bayesian network in Figure (a), which represents a hidden Markov model (HMM). This encodes the joint distribution
P(Q,Y) = P(Q_1) P(Y_1|Q_1) \prod_{t=2}^4 P(Q_t|Q_{t-1}) P(Y_t|Q_t)
For a sequence of length T, we simply ``unroll'' the model for T
time steps. In general, such a dynamic Bayesian network (DBN) can be
specified by just drawing two time slices (this is sometimes called a
2TBN) --- the structure (and parameters) are assumed to repeat.
The Markov property states that the future is independent of the past given the present, i.e., Q_{t+1} \indep Q_{t-1} | Q_t. We can parameterize this Markov chain using a transition matrix, M_{ij} = P(Q_{t+1}=j | Q_t=i), and a prior distribution, \pi_i = P(Q_1 = i).
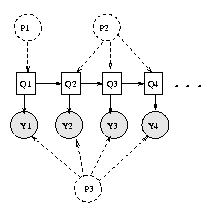
We have assumed that this is a homogeneous Markov chain, i.e., the parameters do not vary with time. This assumption can be made explicit by representing the parameters as nodes: see Figure(b): P1 represents \pi, P2 represents the transition matrix, and P3 represents the parameters for the observation model. If we think of these parameters as random variables (as in the Bayesian approach), parameter estimation becomes equivalent to inference. If we think of the parameters as fixed, but unknown, quantities, parameter estimation requires a separate learning procedure, to be discussed in Section~\ref{sec:learning}. In the latter case, we typically do not represent the parameters in the graph; shared parameters (as in this example) are implemented by specifying that the corresponding CPDs are ``tied''.
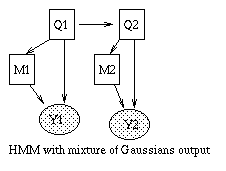
An HMM is a hidden Markov model because we don't see the states of the Markov chain, Q_t, but just a function of them, namely Y_t. For example, if Y_t is a vector, we might define P(Y_t=y|Q_t=i) = N(y; \mu_i, \Sigma_i). A richer model, widely used in speech recognition, is to model the output (conditioned on the hidden state) as a mixture of Gaussians. This is shown below.
|
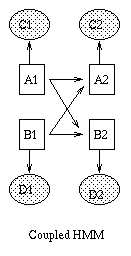
Some popular variations on the basic HMM theme are illustrated below (from left to right: an input-output HMM, a factorial HMM, a coupled HMM). (In the input-output model, the CPD P(Q|U) could be a softmax function, or a neural network.) If we have software to handle inference and learning in general Bayesian networks, all of these models becomes trivial to implement.
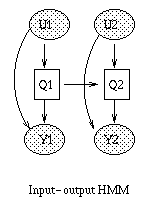
| 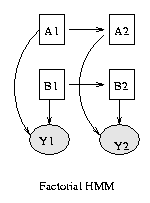
| 
|
A linear dynamical system (LDS) has the same topology as the input-output HMM, except that the hidden nodes have linear-Gaussian CPDs. Replacing Q_t with X_t, the model becomes
P(X_1=x) &=& N(x; x_0, V_0) \\
P(X_{t+1}=x_{t+1}|U_t=u, X_t=x) &=& N(x_{t+1}; A x + B u, Q) \\
P(Y_t=y|X_t=x, U_t=u) &=& N(y; C x + D u, R)
The Kalman filter is an algorithm for computing
P(X_t | y_{1:t}, u_{1:t}) online.
The Rauch-Tung-Striebel (RTS) smoother is an algorithm for computing
P(X_t | y_{1:T}, u_{1:T}) offline.
Both of these algorithms turn out to be special cases of the general
inference algorithms we will discuss in Section~\ref{sec:dynamic_programming}.
|
p(x) = \frac{1}{Z} \prod_{C \in {\cal C}} \psi_C(x_C)
where {\cal C} is the set of maximal cliques in the graph,
\psi_C(x_C) is a potential function (a positive, but otherwise arbitrary,
real-valued function) on the clique x_C, and Z is the
normalization factor
Z = \sum_x \prod_{C \in {\cal C}} \psi_C(x_C).
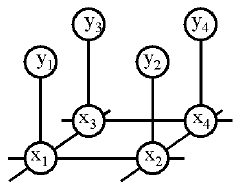
Consider the example above.
In this case, the joint distribution is
P(x,y) \propto \Psi(x_1,x_2) \Psi(x_1,x_3) \Psi(x_2,x_4) \Psi(x_3,x_4)
\prod_{i=1}^4 \Psi(x_i, y_i)
In the Ising model, the graph is a grid, so the cliques are edges, and the X_i nodes are binary. The potentials \Psi(x_i, x_j) for neighboring nodes i,j penalize differences: \Psi(x_i, x_j) = e^{-J(x_i,x_j)}, where J(x_i,x_j) = 1 if x_i=x_j and J(x_i,x_j) = -1 otherwise.
In low-level vision problems (e.g., \cite{Geman84,Freeman00c}), the X_i's are usually hidden, and each X_i node has its own ``private'' observation node Y_i, as in Figure~\ref{fig:vision_mrf}. The potential \Psi(x_i,y_i) = P(y_i|x_i) encodes the local likelihood; this is often a conditional Gaussian, where Y_i is the image intensity of pixel i, and X_i is the underlying (discrete) scene ``label''.
P(X|y) = \frac{P(y|X) P(X)}{P(y)}
where X are the hidden nodes and y is the observed evidence.
(We follow standard practice and represent random {\em variables} by
upper case letters, and sampled {\em values} by lower case letters.)
In words, this formula becomes
conditional likelihood * prior
posterior = ------------------------------
likelihood
In this case, we have
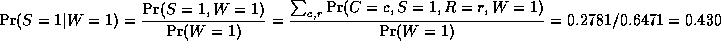
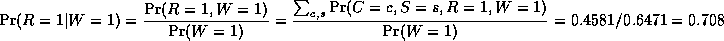
where
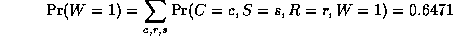
is a normalizing constant, equal to the probability (likelihood) of
the data.
So we see that it is more likely that the grass is wet because
it is raining:
the likelihood ratio is 0.7079/0.4298 = 1.647.
In general, computing posterior esimates using Bayes' rule is computationally intractable. One way to see this is just to consider the normalizing constant, Z: in general, this involves a sum over an exponential number of terms. (For continuous random variables, the sum becomes an integral, which, except for certain notable cases like Gaussians, is not analytically tractable.) We will now see how we can use the conditional independence assumptions encoded in the graph to speed up exact inference.
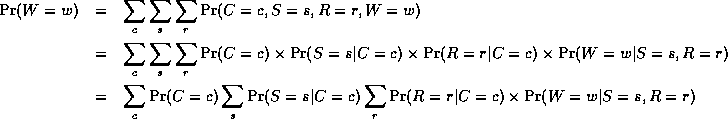
Notice that, as we perform the innermost sums, we create new terms, which need to be summed over in turn e.g.,
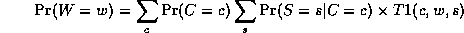
where
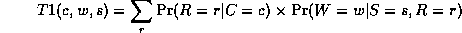
Continuing in this way,
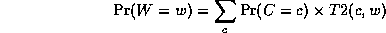
where
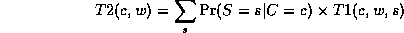
This algorithm is called Variable Elimination. The principle of distributing sums over products can be generalized greatly to apply to any commutative semiring. This forms the basis of many common algorithms, such as Viterbi decoding and the Fast Fourier Transform. For details, see
The amount of work we perform when computing a marginal is bounded by the size of the largest term that we encounter. Choosing a summation (elimination) ordering to minimize this is NP-hard, although greedy algorithms work well in practice.
For details, see
Assuming for now that one is interested in exact inference, the most common approach is therefore to convert the graphical model into a tree, by clustering nodes together (see the example below, from \cite{Russell95}).
Structure Observability Method --------------------------------------------- Known Full Maximum Likelihood Estimation Known Partial EM (or gradient ascent) Unknown Full Search through model space Unknown Partial EM + search through model spaceA third distinction is whether the goal is to find a single ``best'' model/ set of parameters (point estimation), or to return a posterior {\em distribution} over models/ parameters (Bayesian estimation). In the case of known structure, Bayesian parameter estimation is the same as inference, where hidden nodes represent the parameters; this was briefly discussed in Section~\ref{sec:hmm}. Bayesian structure learning will be discussed in Section~\ref{sec:unknown_struct_full_obs_bayes}.
Before going into details, we mention that learning BNs is a huge subject, and this paper only touches the tip of the iceberg. For further details, see the following review papers
L = \frac{1}{M} \log \prod_{m=1}^M \Pr(D_m|G)
= \frac{1}{M} \sum_{i=1}^{n} \sum_{m=1}^M \log P(X_i | Pa(X_i), D_m)
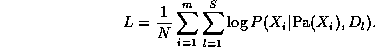
\hat{P}_{ML}(W=w|S=s,R=r) = \frac{\#(W=w,S=s,R=r)}{\#(S=s,R=r)}
where the denominator is \#(S=s,R=r) = \#(W=0,S=s,R=r) + \#(W=1,S=s,R=r).
Thus ``learning'' just amounts to counting (in the case of multinomial
distributions).
If a particular event is not seen in the training set,
it will be assigned a probability of 0.
To avoid this, we can use a Dirichlet prior; this just amounts to
adding ``pseudo counts'' to the empirical counts.
For example, using a uniform Dirichlet prior, the MAP estimate for the
W node becomes
\hat{P}_{MAP}(W=w|S=s,R=r) = \frac{\#(W=w,S=s,R=r) + 1}{\#(S=s,R=r) + 2}
For Gaussian nodes, the MLE of the mean and
covariance are the sample mean and covariance, and
the MLE of the weight matrix is the least squares solution to
the normal equations.
For other kinds of CPDs (e.g., neural networks), more complex procedures are
necessary for computing ML/MAP parameter estimates.
For example, in the case of the W node, we replace the observed counts of the events with the number of times we expect to see each event:
\hat{P}(W=w|S=s,R=r) = E \#(W=w,S=s,R=r) / E \#(S=s,R=r)
where E \#(e) is the expected number of times event e
occurs in the whole training set, given the current guess of the parameters.
These expected counts can be computed as follows
E \#(e) = E \sum_m I(e | D_m) = \sum_m P(e | D_m)where I(e | D_m) is an indicator function which is 1 if event e occurs in training case m, and 0 otherwise. Given the expected counts, we maximize the parameters, and then recompute the expected counts, etc. This iterative procedure is guaranteed to converge to a local maximum of the likelihood surface. (Note: the Baum-Welch algorithm used for training HMMs \cite{Rabiner89} is identical to the EM algorithm.)
It is also possible to do gradient ascent on the likelihood surface (the gradient expression also involves the expected sufficient statistics), but EM is usually faster (since it uses the natural gradient) and simpler (since it has no step size parameter and takes care of parameter constraints automatically. (In the case of multinomials, the constraint is that all the parameters must be in [0,1], and each row of the CPT must sum to one; in the case of Gaussians, the constraint is that the covariance matrix must be positive semi-definite; etc.)
Whether we use EM or gradient ascent, inference becomes a subroutine which is called by the learning procedure; hence fast inference algorithms are crucial for learning.
\Pr(G|D) = \frac{\Pr(D|G) \Pr(G)} {\Pr(D)}
Taking logs, we find
L = \log \Pr(G|D) = \log \Pr(D|G) + \log \Pr(G) + cwhere c = -\log P(D) is a constant independent of G. If the prior probability is higher for simpler models (e.g., ones which are ``sparser'' in some sense), the P(G) term has the effect of penalizing complex models.
In fact, it is not necessary to explicitely penalize complex structures through the structural prior. The marginal likelihood,
P(D|G) = \int P(D|G,\theta) P(\theta|G) d \theta,automatically penalizes more complex structures, because they have more parameters, and hence cannot give as much probability mass to region of space where the data actually lies. (In other words, a complex model is more likely to be ``right'' by chance, and is therefore less believable.) This phenomenon is called Ockham's razor (see e.g., \cite{MacKay95}).
If we assume all the parameters are independent, the marginal likelihood decomposes into a product of local terms:
P(D|G) = \prod_{i=1}^n \int P(X_i | Pa(X_i), \theta_i) P(\theta_i) d \theta_i
In some cases (e.g., if the priors on the parameters are conjugate),
each of these integrals can be performed in closed form, so the
marginal likelihood can be computed very efficiently. (If not, one
typically has to resort to sampling methods.)
The fact that the scoring function is a product of local terms makes local search more efficient, since to compute the relative score of two models that differ by only a few arcs (i.e., neighbors in the space), it is only necessary to compute the terms which they do not have in common --- the other terms cancel when taking the ratio.
P(X|G) = \sum_{Z} \int P(X,Z|G,\theta) P(\theta|G) d \theta
This score does not decompose into a product of local terms.
One approach is to use a Laplace approximation to the posterior of the
parameters \cite{Chickering97}.
After some algebra \cite{Heckerman98}, this leads to the following expression:
\log \Pr(D|G) \approx \log \Pr(D| G, \hat{\theta}_G) - \frac{d}{2} \log M
where M is the number of samples,
\hat{\theta}_G is the ML estimate of the parameters (computed using EM)
and d is the dimension of the model.
(In the fully observable case, the dimension of a model is the number
of free parameters. In a model with hidden variables, it might be less
than this.)
This is called the Bayesian Information Criterion (BIC),
and is equivalent to the Minimum Description Length (MDL) approach.
The first term is just the likelihood and
the second term is a penalty for model complexity. (Note that the BIC
score is independent of the parameter prior.)
Although the BIC score decomposes, local search algorithms (e.g., hill
climbing) are still
expensive, because we need to run EM at each step to compute
\hat{\theta}.
An alternative approach is to do the local search steps
inside of the M step of EM --- this is called Structural EM, and provably converges
to a local maximum of the BIC score \cite{Friedman97}.
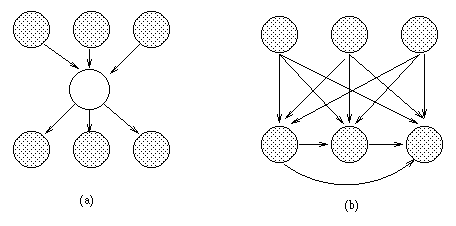
The standard approach is to keep adding hidden nodes one at a time, performing structure learning at each step, until the score drops. There has been some recent work on more intelligent heuristics. For example, dense clique-like graphs (such as that in Figure~\ref{fig:hidden_node}(b)) suggest that a hidden node should be added \cite{Elidan00}.
In {\it sequential} decision theory, the agent (decision maker) is assumed to be interacting with the environment which is modelled as a dynamical system (see Figure~\ref{fig:hmm_variants}(a)). If this dynamical system is linear with Gaussian noise, and the utility function is negative quadratic loss (e.g., consider a missile tracking an airplane, where the goal is to minimize the squared distance between itself and the target), then techniques from control theory can be used to compute the optimal policy, i.e., mapping from percepts to actions. If the system is non-linear, the standard approach is to locally linearize the system.
Linear dynamical systems (LDSs) enjoy the separation property, which states that the optimal behavior can be obtained by first doing state estimation (i.e., infer the hidden states), and then using the expected value of the hidden states as input to a regular LQG (linear quadratic Gaussian) controller. In general, however, the separation property does not hold. For example, consider controlling a mobile robot. The optimal action should take into account the robot's uncertainty about the state of the world (e.g., its location), and not just use the mode of the posterior as if it were the true state of the world. The latter strategy (which is called a certainty-equivalent controller) would never perform information-gathering moves. In other words, the robot might not choose to look before it leapt, no matter how uncertain it was.
In general, finding good controllers for non-linear, partially observed systems, usually with unknown parameters, is extremely challenging. One approach that shows some promise is reinforcement learning. Most of the work has been on systems which are fully observed (e.g., \cite{Sutton98}), but there has been some work on partially observed systems (e.g., \cite{Kaelbling98}). Recent policy search methods (e.g., \cite{Ng00}) show particular promise.
The general framework was developed by Pearl \cite{Pearl88} and various European researchers \cite{Jensen90,Cowell99}, who used it to make probabilistic expert systems. Many of these systems were used for medical diagnosis. A famous example is QMR-DT, a decision-theoretic reformulation of the Quick Medical Reference (QMR) model.
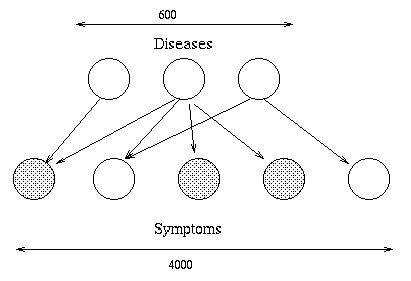
The same kind of diagnosis technology has since been widely adopted by Microsoft, e.g., the Answer Wizard of Office 95, the Office Assistant (the bouncy paperclip guy) of Office 97, and over 30 technical support troubleshooters. In fact, Microsoft now offers a Bayesian network API for Windows developers.
Another interesting fielded application is the Vista system, developed by Eric Horvitz. The Vista system is a decision-theoretic system that has been used at NASA Mission Control Center in Houston for several years. The system uses Bayesian networks to interpret live telemetry and provides advice on the likelihood of alternative failures of the space shuttle's propulsion systems. It also considers time criticality and recommends actions of the highest expected utility. The Vista system also employs decision-theoretic methods for controlling the display of information to dynamically identify the most important information to highlight. Horvitz has gone on to attempt to apply similar technology to Microsoft products, e.g., the Lumiere user interface project \cite{Horvitz98}.
For examples of other applications, see the special issue on UAI of Proc. ACM 38(3), 1995.