Contents
- Set-up repeatable randomness for demo
- Create a Bayes net based on the 'Cancer' topology
- Visualize the ground-truth graph
- Generate a tabular CPD (multinomial model) on each node
- Generate data from the Bayes net
- Use DP algorithm to re-learn the structure
- Use MCMC (Hybrid, Local/Global mixture proposal) to correct the bias induced by modular prior
- Use exhaustive enumeration (sum over super-exponential space of DAGs) to compute exact answer (for a uniform prior)
Set-up repeatable randomness for demo
seed = 0; rand('seed', seed); randn('seed', seed);
Create a Bayes net based on the 'Cancer' topology
labels = {'A','B','C','D','E'};
dag = zeros(5);
A = 1; B = 2; C = 3; D = 4; E = 5;
dag(A,[B C]) = 1;
dag(B,D) = 1;
dag(C,[D E]) = 1;
sz = 2*ones(5,1);
bnet = mk_bnet(dag, sz);
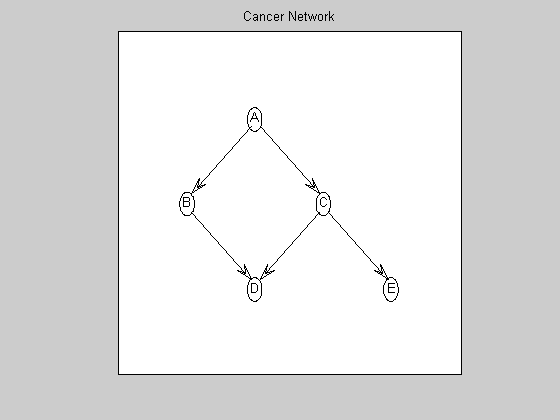
Visualize the ground-truth graph
graphicalLayout = [0.4000 0.2000 0.6000 0.4000 0.8000 ; ... 0.7500 0.5000 0.5000 0.2500 0.2500 ]; % in normalized [0..1]x[0..1] Matlab plotting coordinates figure; myDrawGraph(dag, 'labels', labels, 'layout', graphicalLayout); title('Cancer Network');
Generate a tabular CPD (multinomial model) on each node
p = .9;
cptA = [.4 .6];
cptB = [1-p p ; p 1-p];
cptC = [1-p p ; p 1-p];
cptD = zeros(2, 2, 2);
cptD(:,:,1) = [ .15 .3 ; .7 .9];
cptD(:,:,2) = 1-cptD(:,:,1);
cptE = [p 1-p ; 1-p p];
bnet.CPD{A} = tabular_CPD(bnet, A, 'CPT', cptA);
bnet.CPD{B} = tabular_CPD(bnet, B, 'CPT', cptB);
bnet.CPD{C} = tabular_CPD(bnet, C, 'CPT', cptC);
bnet.CPD{D} = tabular_CPD(bnet, D, 'CPT', cptD);
bnet.CPD{E} = tabular_CPD(bnet, E, 'CPT', cptE);
nNodes = length(dag);
Generate data from the Bayes net
nObservationCases = 350; % # observational data cases nInterventionCases = 0; % no interventions interventions = {}; [data clamped] = mkData(bnet, nObservationCases, interventions, nInterventionCases );
Use DP algorithm to re-learn the structure
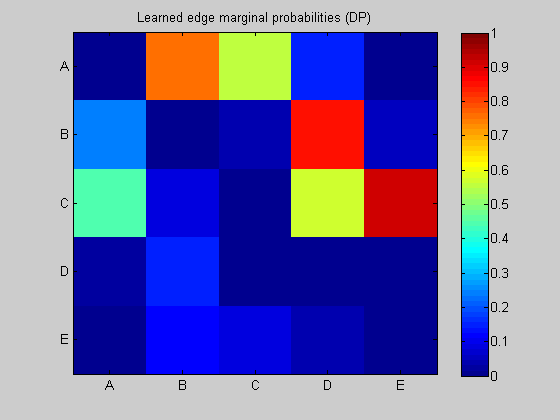
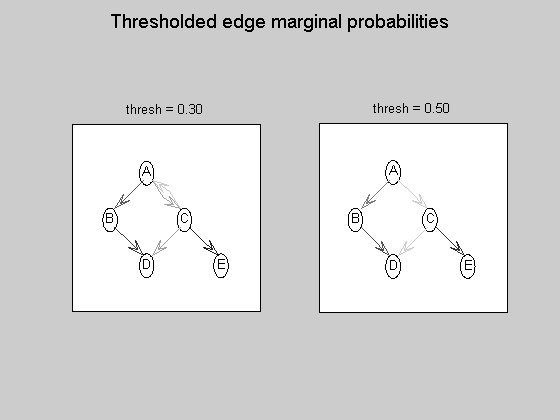
maxFanIn = nNodes - 1; % do not restrict the fan-in of any node aflp = mkAllFamilyLogPrior( nNodes, 'maxFanIn', maxFanIn ); % construct the modular prior on all possible local families aflml = mkAllFamilyLogMargLik( data, 'nodeArity', repmat(2,1,nNodes), 'impossibleFamilyMask', aflp~=-Inf); % compute marginal likelihood on all possible local families ep = computeAllEdgeProb( aflp, aflml ); % compute the marginal edge probabilities using figure; imagesc(ep, [0 1 ]); title('Learned edge marginal probabilities (DP)'); set(gca, 'XTick', 1:5); set(gca, 'YTick', 1:5); set(gca, 'XTickLabel', labels); set(gca, 'YTickLabel', labels); colorbar; figure; subplot(1,2,1); thresh = 0.3; myDrawGraph(ep, 'labels', labels, 'layout', graphicalLayout, 'thresh', thresh); title(sprintf('thresh = %0.2f', thresh)); subplot(1,2,2); thresh = 0.5; myDrawGraph(ep, 'labels', labels, 'layout', graphicalLayout, 'thresh', thresh); title(sprintf('thresh = %0.2f', thresh)); suptitle('Thresholded edge marginal probabilities'); fprintf('Simulation paused. Press any key to continue.'); pause
Simulation paused. Press any key to continue.
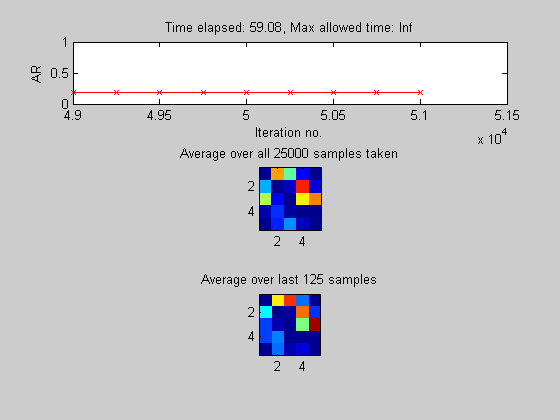
Use MCMC (Hybrid, Local/Global mixture proposal) to correct the bias induced by modular prior
use a uniform prior on structures
[samples, diagnostics] = sampleDags(@uniformGraphLogPrior, aflml, 'burnin', 1000, 'verbose', true, ... 'edgeMarginals', ep, 'koivistoFrac', .1, 'thinning', 2, 'nSamples', 25000); epMcmc = samplesToEdgeMarginals(samples); % average the DAG samples to empirically estimate the edge marginals
Sample 1000 of 51000 [ar=0.198] Sample 2000 of 51000 [ar=0.194] Sample 3000 of 51000 [ar=0.180] Sample 4000 of 51000 [ar=0.191] Sample 5000 of 51000 [ar=0.194] Sample 6000 of 51000 [ar=0.191] Sample 7000 of 51000 [ar=0.188] Sample 8000 of 51000 [ar=0.186] Sample 9000 of 51000 [ar=0.185] Sample 10000 of 51000 [ar=0.186] Sample 11000 of 51000 [ar=0.184] Sample 12000 of 51000 [ar=0.184] Sample 13000 of 51000 [ar=0.184] Sample 14000 of 51000 [ar=0.184] Sample 15000 of 51000 [ar=0.184] Sample 16000 of 51000 [ar=0.184] Sample 17000 of 51000 [ar=0.183] Sample 18000 of 51000 [ar=0.182] Sample 19000 of 51000 [ar=0.182] Sample 20000 of 51000 [ar=0.181] Sample 21000 of 51000 [ar=0.182] Sample 22000 of 51000 [ar=0.182] Sample 23000 of 51000 [ar=0.182] Sample 24000 of 51000 [ar=0.182] Sample 25000 of 51000 [ar=0.180] Sample 26000 of 51000 [ar=0.180] Sample 27000 of 51000 [ar=0.180] Sample 28000 of 51000 [ar=0.181] Sample 29000 of 51000 [ar=0.181] Sample 30000 of 51000 [ar=0.179] Sample 31000 of 51000 [ar=0.179] Sample 32000 of 51000 [ar=0.180] Sample 33000 of 51000 [ar=0.180] Sample 34000 of 51000 [ar=0.181] Sample 35000 of 51000 [ar=0.181] Sample 36000 of 51000 [ar=0.181] Sample 37000 of 51000 [ar=0.182] Sample 38000 of 51000 [ar=0.182] Sample 39000 of 51000 [ar=0.182] Sample 40000 of 51000 [ar=0.182] Sample 41000 of 51000 [ar=0.182] Sample 42000 of 51000 [ar=0.182] Sample 43000 of 51000 [ar=0.182] Sample 44000 of 51000 [ar=0.182] Sample 45000 of 51000 [ar=0.182] Sample 46000 of 51000 [ar=0.182] Sample 47000 of 51000 [ar=0.181] Sample 48000 of 51000 [ar=0.180] Sample 49000 of 51000 [ar=0.181] Sample 50000 of 51000 [ar=0.181] Sample 51000 of 51000 [ar=0.182]
Use exhaustive enumeration (sum over super-exponential space of DAGs) to compute exact answer (for a uniform prior)
(DP algorithm is exact too, but only for the restricted "modular" prior)
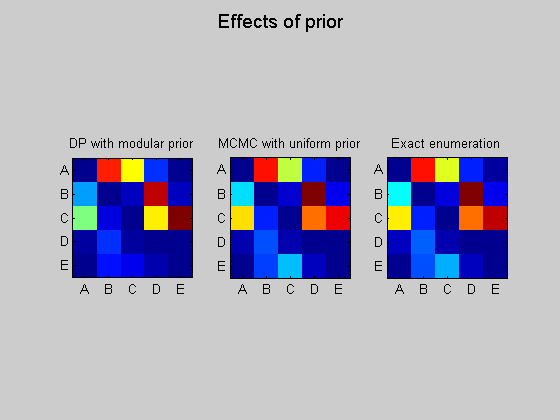
epExact = computeAllEdgeProb_Exact(0, aflml); % 0 denotes uniform prior figure; subplot(1,3,1); imagesc(ep); set(gca, 'XTick', 1:5); set(gca, 'YTick', 1:5); set(gca, 'XTickLabel', labels); set(gca, 'YTickLabel', labels); title('DP with modular prior'); axis('square'); subplot(1,3,2); imagesc(epMcmc); set(gca, 'XTick', 1:5); set(gca, 'YTick', 1:5); set(gca, 'XTickLabel', labels); set(gca, 'YTickLabel', labels); title('MCMC with uniform prior'); axis('square'); subplot(1,3,3); imagesc(epExact); set(gca, 'XTick', 1:5); set(gca, 'YTick', 1:5); set(gca, 'XTickLabel', labels); set(gca, 'YTickLabel', labels); title('Exact enumeration'); axis('square'); suptitle('Effects of prior');
loading Cache/DAGS5.mat likelihood 10000/29281 likelihood 20000/29281 marginals 10000/29281 marginals 20000/29281