NOTE: CPSC 545 students are only required to solve problems 1 and 4; CPSC 445 students are expected to solve all problems.
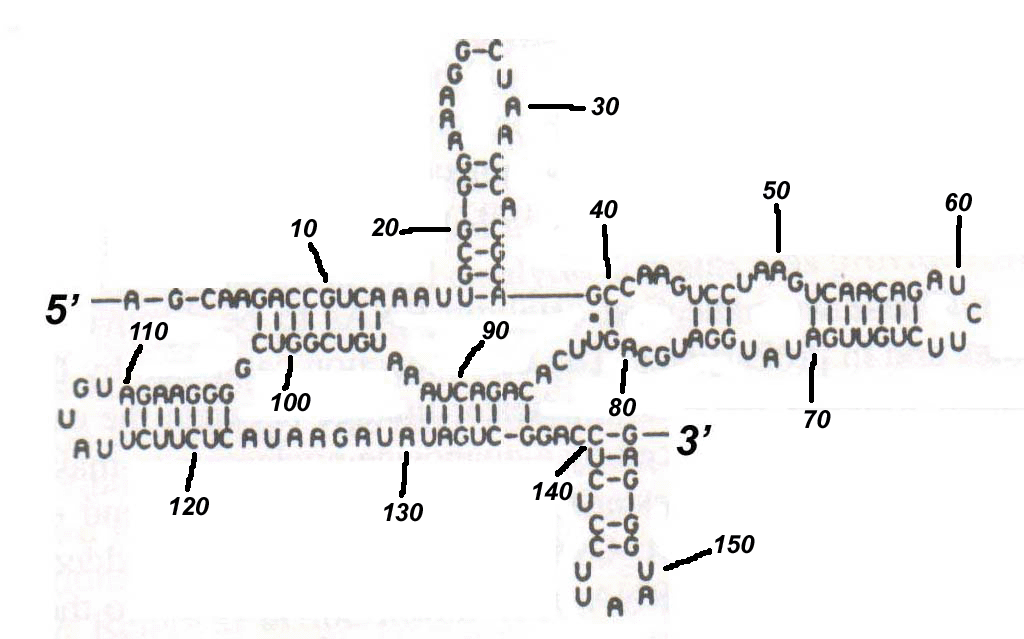
(a) Given a figure (Figure 1) of the RNA molecule, name all of the secondary structure elements, and give their base pair position.
[2
marks]

(b) Make structure from part (a) pseudoknot free by removing minimal number of bases. [1 marks]
(c) Calculate energy of the structure given in Figure 2, based on the Zuker's energy parameters that can be found at http://www.bioinfo.rpi.edu/~zukerm/rna/energy/, use tables for 37 degree Celsius for RNA folding. [4 marks]
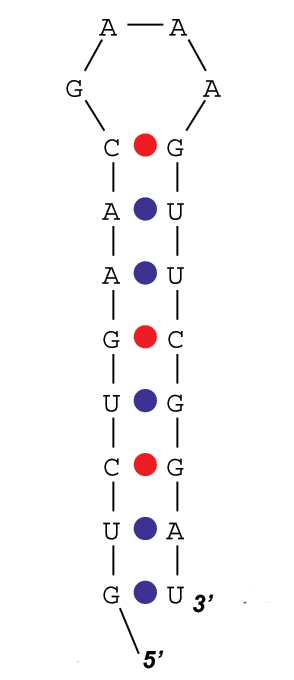
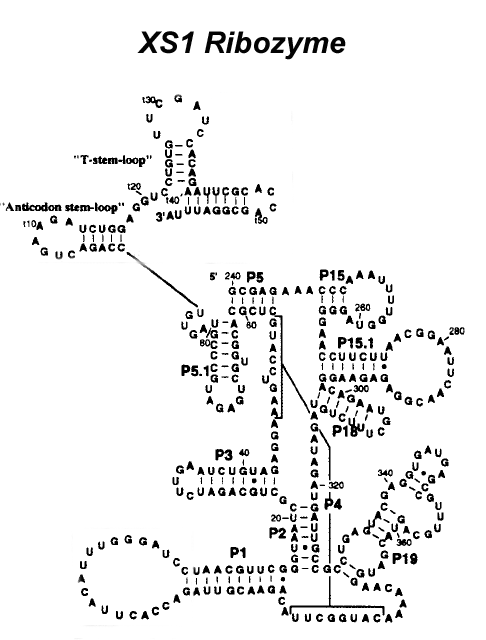
(a) Given a sequence of biological pseudoknotted XS1 rybozyme, use
MFOLD program of Zucker et al. to predict secondary structure, and compare optimal
structure to the original biological structure given in Figure 3. What do you notice? RNA sequence is:
GCGAGAAACCCAAAUUUUGGUAGGGGAACCUUCUUAACGGAAUUC
AACGGAGAGAAGGACAGAAUGCUUUCUGUAGAUAGAUGAUUGCCG
CCUGAGUACGAGGUGAUGAGCCGUUUGCAGUACGAUGGAACAAAA
CAUGGCUUACAGAACGUUAGACCACUUACAUUUGGGAUCCUAACG
UUCGGGUAAUCGCUGCAGAUCUUGAAUCUGUAGAGGAAAGUCCAU
GCUCGCACGGUGCUGAGAUGCCCGUAGUGUCCAGACUGAAGAUCU
GGAGGUCCUGUGUUCGAUCCACAGAAUUCGCACCAGCGGAUUUA
[2
marks]
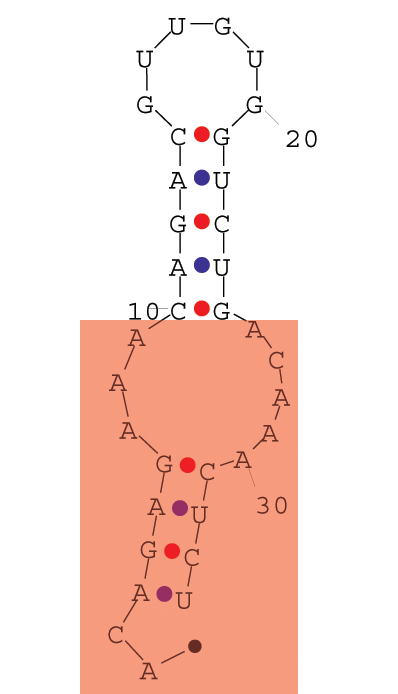
(b) Given a sequence of a small RNA structure depicted in figure, design a small piece of RNA that when both sequences are inputed in PairFold program of Andronescu et. al, the stem and the internal loop marked in red in the Figure 4 will dissapear. RNA sequence is: ACAGAGAAACAGACGUUGUGGUCUGACAAACUCU [2 marks]
(c) [3 marks]
(i) Design a sequence that folds into the structure given in the parenthesis notation:
((((((.((.(((((((((.......(((...))).........)))))..))))..)))))))).
using DESIGNER program of Andronescu et. al.
(ii) Fold the original biological sequence:
AAACAGAGAAGUCAACCAGAGAAACAGACGUUGUCGUAUAAUACCUGGUACUGACAGUCCUGUUUU
using MFOLD.
(iii) Comment on the energies of structures obtained by MFOLD and the
energy calculated by the RNA DESIGNER.
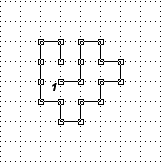
Given a protein structure on the 2 dimensional lattice under Hydrophobic Polar (HP) model, design a sequence that folds into the target structure producing minimal free energy and has a maximum of 10 hydrophobic (h) amino acids, specify both the designed sequence and its minimal free energy. [3 marks]
(a) Explain when threading and when homology is an appropriate method for protein structure prediction. [2 marks]
(b) Explain the difference between protein fold and protein domain (cite any sources where appropriate). [1 marks]
(c) Explain the difference between secondary and super-secondary protein structure. [1 marks]
(d) List three reasons why normalization is performed on microarray data before analysis. [3 marks]
(e) Imagine that you decided to test gene expression in normal and mutant mice, you carried out microarray experiment, extracted gene expression data, and made a scatter plot of relative expression for all genes (of normal versus mutant condition), what do you expect the scatter plot looks like knowing that only few genes are differentially expressed? [3 marks]