CPSC536A - Notes for Class 3
11.1.2001
Index
1. Mutations (cont.)
2. Tree of Life
3. Techniques in Molecular Biology
4. Topics in Bioinformatics
1. Mutations (cont.)
In addition to the class of mutations mentioned in section 8 of last week's
notes, the following 2 classes exist.
1.2 Insertions and Deletions
Insertion mutations introduce large sequences of DNA into the
genome, while Deletion mutations remove large quantities. If the
number of base pairs added/removed is not a multiple of 3, this
mutation is (in addition) a frame shift mutation. Retro viruses
can cause insertions.
(see slide #1)
1.3 Crossover
Crossover mutations occur in a diploid organism (ie. 2
sets of homologous chromosomes) when a DNA part of the first
chromosome is built into the second chromosome and vice versa. One
DNA of the first chromosome continues on the second chromosome and
replaces one DNA there. This DNA crosses over to the other DNA of
the first chromosome and completes it. The recombination can lead to
huge sequence changes in the genome.
(see slide #2)
2. Tree of Life
The Tree of Life (Phylogenetic Tree) charts a trace-back for all
organisms to either the ancestral prokaryote (for
bacteria and similar) or the ancestral eukaryote (for
higher organisms) (see slide #3). For more on prokaryotes
vs. eukaryotes read section 4 of last
week's notes. Mitochondria and chloroplasts may have developed
as symbionts in prokaryotic organisms (endosymbiont
hypothesis). (see slide #4) The internal nodes of the tree
are hypothetical and do not necessarily correspond to actual organisms. They
represent common differences in the DNA along a path in the
tree. The early development (i.e. close to the root) along this
tree may be due to simple mutations, while later development
involves more recombination mutations (insert &
delete, crossover).
3. Techniques in Molecular Biology
3.1 Restriction
The Restriction technique uses Restriction Enzymes to cut
DNA at a particular recognition sequence (of only a few
base pairs). Not every possible short sequence has a corresponding
Restriction Enzyme. Some Restriction Enzymes create blunt
ends (eg. Hpa I), others create sticky or
staggered ends (eg. Eco RI) (see slide#7). Notice that
Mbo II does not cut the DNA at its recognition sequence. (see
slide #8)
In nature, Restriction Enzymes play an important role in chopping
up DNA during "import" of outside DNA. Engineers chop up DNA for
further analysis.
3.2 Electrophoresis
Electrophoresis is a technique to separate and measure the lengths
of a mix of DNA, RNA, or proteins. An electric field is used to drag the
molecules along a gel (the shorter the molecule, the less
resistance in the gel, the further the molecule advances in the
gel). (see slides #9, #10)
3.3 Gel-Transfer Hybridization
Gel-Transfer Hybridization is used to detect complementary
sequences, eg. in search for exon/intron. First the DNA is split
into 2 single strands (denaturing). Then a corresponding
mRNA is added and hybridizes with a DNA single strand to a double
strand. The bend out intron sequence is removed, the double strand
is denatured, and the exons can be detected by
electrophoresis. (see slide #13)
Techniques similar to this technique (southern
blotting) are northern blotting (for RNA) and
western blotting (for proteins).
PCR
The Polymerase Chain Reaction (PCR) is used to get big amounts of
identical DNA used for further analysis. It is a technically cheap
method of in vitro DNA replication. It starts with denaturing the
"original" DNA piece (not shown on slide #15). A build-to-order
primer (of ~20 bases in length known to be part of the sequence)
is added. DNA polymerase starts to build the second strand
starting at the primer. When this is done, the double strand is
denatured again, and the cycle restarts (see slides #16,#17). The
reaction uses up nucleotides and primers, while the enzymes
involved (in particular, DNA polymerase) survive the process.
Sequencing
Sequencing is the method of determining the sequence of DNA. The
Sänger method (see slide #18) is based on provoked chain
termination. A single strand of to be sequenced DNA gets completed
(oligonucleotide primer,...) from a pool of nucleotides. In the
first reaction, a small amount of one nucleotide (eg. A) in the pool
is altered in a way that it will terminate the replication when it
is used. In a massive parallel process, these marked nucleotides
will be eventually used on any corresponding position of the
sequence. The length of all this partial replicas ending in A can
be measured at single base pair precision (Electrophoresis). Three analogous reactions
are performed for the other nucleotides and yield lengths as well. The
DNA sequence is given by the names of the last nucleotide of the
replicas in order of increasing length. The example at the bottom
of slide #18 shows the sequence ATGTCAGTCCAG.
The process of building partial replicas and reading the
lengths can be automated. The altered nucleotides are distinctly
marked, and the 4 types of partial replicas are build in one
process. The replicas "race" in one gel-lane and the sequence is
determined by laser-reading the (noisy) marks (coded as colors)
of the nucleotides along the lane. (see slides #19, #20)
Cloning
Originally, cloning referred to creating a replica of something,
while it now is also used in a different sense. One example was
described (slide #21).
Small DNA rings (plasmids) are opened using
Restriction. A new piece of DNA is annealed to the two sticky
ends. The result is a new, altered plasmid as well as unwanted
n- (original), 2n-, 3n, ...-sized rings.
The construct can be amplified or translated into proteins
in the bacterium E. Coli; there are also techniques for testing whether
the insertion was successful. Slides #22, #23: The plasmid
contains a residence gene (AmpR), so
that the presence of the plasmid in E. Coli can be determined by
checking for this resistance. If LacZ' is inoperative, the
insertion was successful.
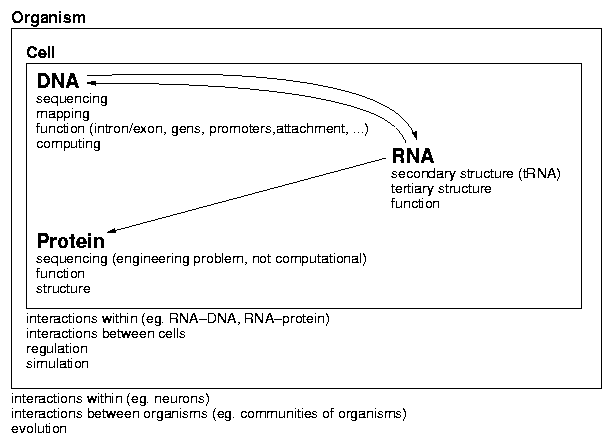
Topics in Bioinformatics
$Id: class3-notes.html,v 1.8 2001/01/16 06:59:49 ksjuse Exp $
Kai S. Juse
Last modified: Tue Jan 16 12:31:30 PST 2001
