CS536A - Class Notes 01/03/08
Module 5 - GENE FINDING
- increasing amount of genomic data available, but interpretation lags
behind
Problems:
-
intron/exon boundaries
-
lots of noncoding regions - only ~3% is coding/exons
-
alternative splicing - 35% of genes
-
overlapping genes
-
pseudogenes
-
regulatory regions (TATA, CAAT boxes) are important for gene expression,
but location with respect to genes are not uniquely determined.
-
Basic regulatory elements are usually upstream of transcription start site,
but enhancers, silencers can be upstream, downstream, and in introns.
- these problems closely related to fundamental issuesin transcription,
translation, and splicing(RNA)
Computational Gene Finding
given raw sequence (DNA), predict:
-
coding/noncoding regions
-
introns/exons
-
splicing patterns
-
transcription factor binding sites
Naive approach
search for characteristic subseqs (eg. GT----AG at intron exon boundaries,
TATA, etc.) by pattern matching
Problem
-
in this approach, we only find the most conserved signals
-
not sufficient to characterize genes/exons
Ideal Approach
completely simulate transcription, splicing and translation
Problem
simulation would be too complex, even IF we knew everything
Simplified Approach (in prokaryotes only)
-
no introns, so look for long uninterrupted open reading frames (ORFs)
-
BUT only FEW genes like this in eukaryotes and distribution of ORF lengths
(in humans) seems totally random.
How it's really done
-
signal detection (splice sites, promoters, etc.)
-
compositional properties of coding vs. noncoding regions (GC content, hexamer
frequency)
-
OR, better yet, integration of these methods with homology search
-
modern gene finders predict individual functional elements as well as complete
gene structure (i.e. the set of spliceable introns)
Signal Detection
-
simple motif search not good enough
-
Weight Matrix Method (WMM)
ASIDE: Shannon information content 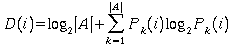
where
A is the alphabet {A, C, G, T}
|A| is 4
Pk(i) is the probability of observing base k in position
i
so for random sequence P = 1/4 and D(i) = 0 = 2 + 1/4 log2(1/4)
+ 1/4 log2(1/4) + 1/4 log2(1/4) + 1/4 log2(1/4)
This score is a bit score and gives a figure for the information conferred
by a given base being at the given position.
this gives rise to sequence logos with height proporitonal to this bit
score, looking like:
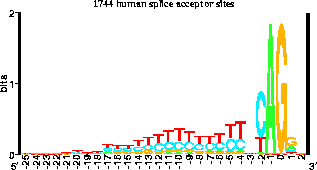
(From http://www.lecb.ncifcrf.gov/~toms/introduction.html)
Weight Matrix example for a splice donor site
Position-> 1 2 3 4 5
6 Multiply A C G T
...to get
A
0 0 0 1 1 0 this
1 1
an additive
C
0 0 0 0 0 0 by the 2
1 score.
G
1 10 0 1 1 1 "data 3
1
T
0 0 10 0 0 0 matrix" 4
1
5 1
6 1
More rigorous model - probablistic WMM version
given frequencies Pk(i) of nucleotide k at position i
and sequence X = X1...Xn
probability of generating X = PX1(1)*PX2(2)*.....*PXn(n)
Generalization
Weight Array Method (WAM) - these model pairwise dependencies of positions
(e.g. in RNA secondary structure)
Where do model parameters come from?
-
training: "learning from aligned sequence data of signals
-
manual determination (relies on human experience)
