GEViTRec: Data Reconnaissance Through Recommendation Using a Domain-Specific Visualization Prevalence Design Space
Abstract |
Paper |
Open Source |
Figures |
Supplemental Material
Abstract
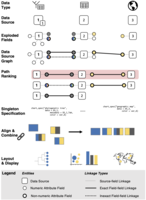
Genomic Epidemiology (genEpi) is a branch of public health that uses many different data types including tabular, network, genomic, and geographic, to identify and contain outbreaks of deadly diseases. Due to the volume and variety of data, it is challenging for genEpi domain experts to conduct data reconnaissance; that is, have an overview of the data they have and make assessments toward its quality, completeness, and suitability. We present an algorithm for data reconnaissance through automatic visualization recommendation, GEViTRec. Our approach handles a broad variety of dataset types and automatically generates visually coherent combinations of charts, in contrast to existing systems that primarily focus on singleton visual encodings of tabular datasets. We automatically detect linkages across multiple input datasets by analyzing non-numeric attribute fields, creating a data source graph within which we analyze and rank paths. For each high-ranking path, we specify chart combinations with positional and color alignments between shared fields, using a gradual binding approach to transform initial partial specifications of singleton charts to complete specifications that are aligned and oriented consistently. A novel aspect of our approach is its combination of domain-agnostic elements with domain-specific information that is captured through a domain-specific visualization prevalence design space. Our implementation is applied to both synthetic data and real Ebola outbreak data. We compare GEViTRec's output to what previous visualization recommendation systems would generate, and to manually crafted visualizations used by practitioners. We conducted formative evaluations with ten genEpi experts to assess the relevance and interpretability of our results.
Paper
GEViTRec: Data Reconnaissance Through Recommendation Using a Domain-Specific Visualization Prevalence Design Space
Open Source
»
Open source code on Github
High-Resolution Figures
Supplementary Materials
»
Supplemental Material (PDF)
- S1 Implementation Details
- S2 Survey Procedures
- S3 Survey Demos: Synthetic Data and Ebola Outbreak data
- S4 Survey Results
- S5 Survey Administrator Notes
- S6 Survey Session Partial Transcripts
- S7 Comparison to Previous Work
Last modified: Mon Oct 25 14:12:44 PDT 2021